| Reaction Details |
|---|
 | Report a problem with these data |
| Target | RecBCD enzyme subunit RecD |
|---|
| Ligand | BDBM57102 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Counterscreen for AddAB inhibitors: absorbance-based bacterial cell-based high throughput dose response assay to identify inhibitors of RecBCD |
|---|
| IC50 | >39496±n/a nM |
|---|
| Citation |  PubChem, PC Counterscreen for AddAB inhibitors: absorbance-based bacterial cell-based high throughput dose response assay to identify inhibitors of RecBCD PubChem Bioassay(2010)[AID] PubChem, PC Counterscreen for AddAB inhibitors: absorbance-based bacterial cell-based high throughput dose response assay to identify inhibitors of RecBCD PubChem Bioassay(2010)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| RecBCD enzyme subunit RecD |
|---|
| Name: | RecBCD enzyme subunit RecD |
|---|
| Synonyms: | RECD_ECOLI | exonuclease V (RecBCD complex), alpha chain | hopE | recD |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 66906.68 |
|---|
| Organism: | Escherichia coli str. K-12 substr. MG1655 |
|---|
| Description: | gi_16130723 |
|---|
| Residue: | 608 |
|---|
| Sequence: | MKLQKQLLEAVEHKQLRPLDVQFALTVAGDEHPAVTLAAALLSHDAGEGHVCLPLSRLEN
NEASHPLLATCVSEIGELQNWEECLLASQAVSRGDEPTPMILCGDRLYLNRMWCNERTVA
RFFNEVNHAIEVDEALLAQTLDKLFPVSDEINWQKVAAAVALTRRISVISGGPGTGKTTT
VAKLLAALIQMADGERCRIRLAAPTGKAAARLTESLGKALRQLPLTDEQKKRIPEDASTL
HRLLGAQPGSQRLRHHAGNPLHLDVLVVDEASMIDLPMMSRLIDALPDHARVIFLGDRDQ
LASVEAGAVLGDICAYANAGFTAERARQLSRLTGTHVPAGTGTEAASLRDSLCLLQKSYR
FGSDSGIGQLAAAINRGDKTAVKTVFQQDFTDIEKRLLQSGEDYIAMLEEALAGYGRYLD
LLQARAEPDLIIQAFNEYQLLCALREGPFGVAGLNERIEQFMQQKRKIHRHPHSRWYEGR
PVMIARNDSALGLFNGDIGIALDRGQGTRVWFAMPDGNIKSVQPSRLPEHETTWAMTVHK
SQGSEFDHAALILPSQRTPVVTRELVYTAVTRARRRLSLYADERILSAAIATRTERRSGL
AALFSSRE
|
|
|
|---|
| BDBM57102 |
|---|
| n/a |
|---|
| Name | BDBM57102 |
|---|
| Synonyms: | MLS000541562 | N-cyclohexyl-6-phenyl-3-(2-pyridinyl)-1,2,4-triazin-5-amine | N-cyclohexyl-6-phenyl-3-pyridin-2-yl-1,2,4-triazin-5-amine | SMR000126420 | cid_6404647 | cyclohexyl-[6-phenyl-3-(2-pyridyl)-1,2,4-triazin-5-yl]amine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H21N5 |
|---|
| Mol. Mass. | 331.4142 |
|---|
| SMILES | C1CCC(CC1)Nc1nc(nnc1-c1ccccc1)-c1ccccn1 |
|---|
| Structure | 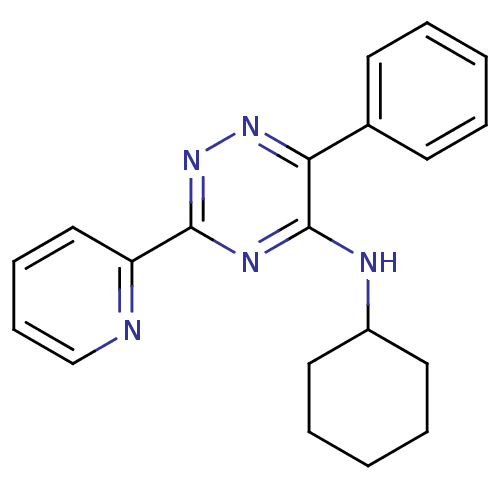
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Counterscreen for AddAB inhibitors: absorbance-based bacterial cell-based high throughput dose response assay to identify inhibitors of RecBCD PubChem Bioassay(2010)[AID]
PubChem, PC Counterscreen for AddAB inhibitors: absorbance-based bacterial cell-based high throughput dose response assay to identify inhibitors of RecBCD PubChem Bioassay(2010)[AID]