| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Gamma-aminobutyric acid receptor subunit alpha-1 |
|---|
| Ligand | BDBM50056101 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | >10000±n/a nM |
|---|
| Comments | PDSP_2202 |
|---|
| Citation |  Nishida, A; Miyata, K; Tsutsumi, R; Yuki, H; Akuzawa, S; Kobayashi, A; Kamato, T; Ito, H; Yamano, M; Katuyama, Y Pharmacological profile of (R)-1-[2,3-dihydro-1-(2'-methylphenacyl)-2-oxo- 5-phenyl-1H-1,4-benzodiazepin-3-yl]-3-(3-methylphenyl)urea (YM022), a new potent and selective gastrin/cholecystokinin-B receptor antagonist, in vitro and in vivo. J Pharmacol Exp Ther269:725-31 (1994) [PubMed] Nishida, A; Miyata, K; Tsutsumi, R; Yuki, H; Akuzawa, S; Kobayashi, A; Kamato, T; Ito, H; Yamano, M; Katuyama, Y Pharmacological profile of (R)-1-[2,3-dihydro-1-(2'-methylphenacyl)-2-oxo- 5-phenyl-1H-1,4-benzodiazepin-3-yl]-3-(3-methylphenyl)urea (YM022), a new potent and selective gastrin/cholecystokinin-B receptor antagonist, in vitro and in vivo. J Pharmacol Exp Ther269:725-31 (1994) [PubMed] |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Gamma-aminobutyric acid receptor subunit alpha-1 |
|---|
| Name: | Gamma-aminobutyric acid receptor subunit alpha-1 |
|---|
| Synonyms: | Benzodiazepine central | Benzodiazepine receptors | GABA A Benzodiazepine | GABA A Benzodiazepine Type I | GABA A Benzodiazepine Type II | GABA A Benzodiazepine Type IIL | GABA A Benzodiazepine Type IIM | GABA A Benzodiazepine omega1 | GABA A Benzodiazepine omega2 | GABA A Benzodiazepine omega5 | GABA A alpha1 | GABA A anti-Alpha1 | GABA receptor alpha-1 subunit | GABA, Chloride, TBOB | GABA-PICROTOXIN | GBRA1_RAT | Gabra-1 | Gabra1 | TBPS |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 51770.21 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P62813 |
|---|
| Residue: | 455 |
|---|
| Sequence: | MKKSRGLSDYLWAWTLILSTLSGRSYGQPSQDELKDNTTVFTRILDRLLDGYDNRLRPGL
GERVTEVKTDIFVTSFGPVSDHDMEYTIDVFFRQSWKDERLKFKGPMTVLRLNNLMASKI
WTPDTFFHNGKKSVAHNMTMPNKLLRITEDGTLLYTMRLTVRAECPMHLEDFPMDAHACP
LKFGSYAYTRAEVVYEWTREPARSVVVAEDGSRLNQYDLLGQTVDSGIVQSSTGEYVVMT
THFHLKRKIGYFVIQTYLPCIMTVILSQVSFWLNRESVPARTVFGVTTVLTMTTLSISAR
NSLPKVAYATAMDWFIAVCYAFVFSALIEFATVNYFTKRGYAWDGKSVVPEKPKKVKDPL
IKKNNTYAPTATSYTPNLARGDPGLATIAKSATIEPKEVKPETKPPEPKKTFNSVSKIDR
LSRIAFPLLFGIFNLVYWATYLNREPQLKAPTPHQ
|
|
|
|---|
| BDBM50056101 |
|---|
| n/a |
|---|
| Name | BDBM50056101 |
|---|
| Synonyms: | 1-[(R)-2-Oxo-1-(2-oxo-2-o-tolyl-ethyl)-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl]-3-m-tolyl-urea | CHEMBL115121 | YM022 | YM022 Racemate |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C32H28N4O3 |
|---|
| Mol. Mass. | 516.5897 |
|---|
| SMILES | Cc1cccc(NC(=O)N[C@@H]2N=C(c3ccccc3)c3ccccc3N(CC(=O)c3ccccc3C)C2=O)c1 |t:11| |
|---|
| Structure | 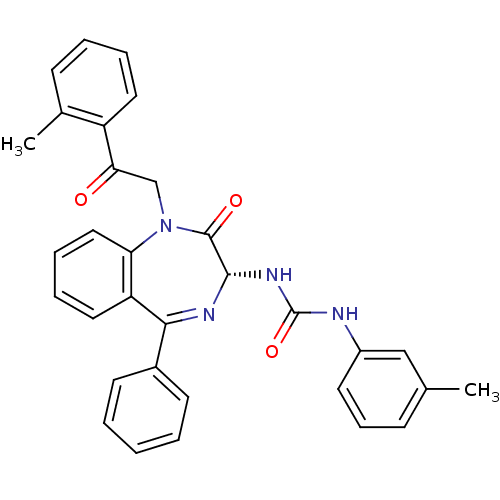
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Nishida, A; Miyata, K; Tsutsumi, R; Yuki, H; Akuzawa, S; Kobayashi, A; Kamato, T; Ito, H; Yamano, M; Katuyama, Y Pharmacological profile of (R)-1-[2,3-dihydro-1-(2'-methylphenacyl)-2-oxo- 5-phenyl-1H-1,4-benzodiazepin-3-yl]-3-(3-methylphenyl)urea (YM022), a new potent and selective gastrin/cholecystokinin-B receptor antagonist, in vitro and in vivo. J Pharmacol Exp Ther269:725-31 (1994) [PubMed]
Nishida, A; Miyata, K; Tsutsumi, R; Yuki, H; Akuzawa, S; Kobayashi, A; Kamato, T; Ito, H; Yamano, M; Katuyama, Y Pharmacological profile of (R)-1-[2,3-dihydro-1-(2'-methylphenacyl)-2-oxo- 5-phenyl-1H-1,4-benzodiazepin-3-yl]-3-(3-methylphenyl)urea (YM022), a new potent and selective gastrin/cholecystokinin-B receptor antagonist, in vitro and in vivo. J Pharmacol Exp Ther269:725-31 (1994) [PubMed]