| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Receptor-interacting serine/threonine-protein kinase 1 |
|---|
| Ligand | BDBM15003 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1673964 (CHEMBL4023993) |
|---|
| Ki | 2.0±n/a nM |
|---|
| Citation |  Yoshikawa, M; Saitoh, M; Katoh, T; Seki, T; Bigi, SV; Shimizu, Y; Ishii, T; Okai, T; Kuno, M; Hattori, H; Watanabe, E; Saikatendu, KS; Zou, H; Nakakariya, M; Tatamiya, T; Nakada, Y; Yogo, T Discovery of 7-Oxo-2,4,5,7-tetrahydro-6 H-pyrazolo[3,4- c]pyridine Derivatives as Potent, Orally Available, and Brain-Penetrating Receptor Interacting Protein 1 (RIP1) Kinase Inhibitors: Analysis of Structure-Kinetic Relationships. J Med Chem61:2384-2409 (2018) [PubMed] Article Yoshikawa, M; Saitoh, M; Katoh, T; Seki, T; Bigi, SV; Shimizu, Y; Ishii, T; Okai, T; Kuno, M; Hattori, H; Watanabe, E; Saikatendu, KS; Zou, H; Nakakariya, M; Tatamiya, T; Nakada, Y; Yogo, T Discovery of 7-Oxo-2,4,5,7-tetrahydro-6 H-pyrazolo[3,4- c]pyridine Derivatives as Potent, Orally Available, and Brain-Penetrating Receptor Interacting Protein 1 (RIP1) Kinase Inhibitors: Analysis of Structure-Kinetic Relationships. J Med Chem61:2384-2409 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Receptor-interacting serine/threonine-protein kinase 1 |
|---|
| Name: | Receptor-interacting serine/threonine-protein kinase 1 |
|---|
| Synonyms: | Cell death protein RIP | RIP | RIP-1 | RIP1 | RIPK1 | RIPK1_HUMAN | Receptor-interacting protein 1 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 75926.99 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q13546 |
|---|
| Residue: | 671 |
|---|
| Sequence: | MQPDMSLNVIKMKSSDFLESAELDSGGFGKVSLCFHRTQGLMIMKTVYKGPNCIEHNEAL
LEEAKMMNRLRHSRVVKLLGVIIEEGKYSLVMEYMEKGNLMHVLKAEMSTPLSVKGRIIL
EIIEGMCYLHGKGVIHKDLKPENILVDNDFHIKIADLGLASFKMWSKLNNEEHNELREVD
GTAKKNGGTLYYMAPEHLNDVNAKPTEKSDVYSFAVVLWAIFANKEPYENAICEQQLIMC
IKSGNRPDVDDITEYCPREIISLMKLCWEANPEARPTFPGIEEKFRPFYLSQLEESVEED
VKSLKKEYSNENAVVKRMQSLQLDCVAVPSSRSNSATEQPGSLHSSQGLGMGPVEESWFA
PSLEHPQEENEPSLQSKLQDEANYHLYGSRMDRQTKQQPRQNVAYNREEERRRRVSHDPF
AQQRPYENFQNTEGKGTAYSSAASHGNAVHQPSGLTSQPQVLYQNNGLYSSHGFGTRPLD
PGTAGPRVWYRPIPSHMPSLHNIPVPETNYLGNTPTMPFSSLPPTDESIKYTIYNSTGIQ
IGAYNYMEIGGTSSSLLDSTNTNFKEEPAAKYQAIFDNTTSLTDKHLDPIRENLGKHWKN
CARKLGFTQSQIDEIDHDYERDGLKEKVYQMLQKWVMREGIKGATVGKLAQALHQCSRID
LLSSLIYVSQN
|
|
|
|---|
| BDBM15003 |
|---|
| n/a |
|---|
| Name | BDBM15003 |
|---|
| Synonyms: | 3-(4-{4-aminofuro[2,3-d]pyrimidin-5-yl}phenyl)-1-[2-fluoro-5-(trifluoromethyl)phenyl]urea | 4-amino-5-diarylurea-furo[2,3-d]pyrimidine 8a | D3RKN_27 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H13F4N5O2 |
|---|
| Mol. Mass. | 431.3431 |
|---|
| SMILES | Nc1ncnc2occ(-c3ccc(NC(=O)Nc4cc(ccc4F)C(F)(F)F)cc3)c12 |
|---|
| Structure | 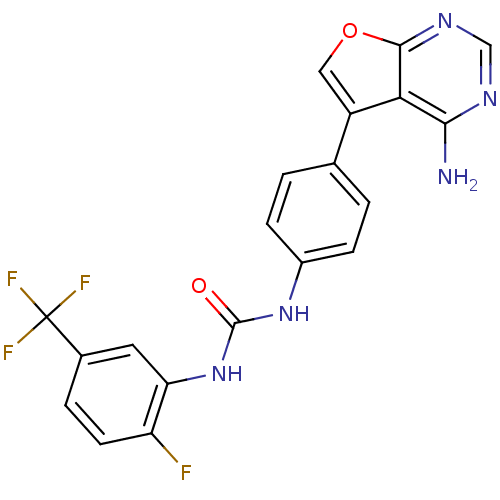
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Yoshikawa, M; Saitoh, M; Katoh, T; Seki, T; Bigi, SV; Shimizu, Y; Ishii, T; Okai, T; Kuno, M; Hattori, H; Watanabe, E; Saikatendu, KS; Zou, H; Nakakariya, M; Tatamiya, T; Nakada, Y; Yogo, T Discovery of 7-Oxo-2,4,5,7-tetrahydro-6 H-pyrazolo[3,4- c]pyridine Derivatives as Potent, Orally Available, and Brain-Penetrating Receptor Interacting Protein 1 (RIP1) Kinase Inhibitors: Analysis of Structure-Kinetic Relationships. J Med Chem61:2384-2409 (2018) [PubMed] Article
Yoshikawa, M; Saitoh, M; Katoh, T; Seki, T; Bigi, SV; Shimizu, Y; Ishii, T; Okai, T; Kuno, M; Hattori, H; Watanabe, E; Saikatendu, KS; Zou, H; Nakakariya, M; Tatamiya, T; Nakada, Y; Yogo, T Discovery of 7-Oxo-2,4,5,7-tetrahydro-6 H-pyrazolo[3,4- c]pyridine Derivatives as Potent, Orally Available, and Brain-Penetrating Receptor Interacting Protein 1 (RIP1) Kinase Inhibitors: Analysis of Structure-Kinetic Relationships. J Med Chem61:2384-2409 (2018) [PubMed] Article