| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 |
|---|
| Ligand | BDBM50250859 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_1682355 |
|---|
| IC50 | 0.670000±n/a nM |
|---|
| Citation |  Butler, CR; Beck, EM; Harris, A; Huang, Z; McAllister, LA; Am Ende, CW; Fennell, K; Foley, TL; Fonseca, K; Hawrylik, SJ; Johnson, DS; Knafels, JD; Mente, S; Noell, GS; Pandit, J; Phillips, TB; Piro, JR; Rogers, BN; Samad, TA; Wang, J; Wan, S; Brodney, MA Azetidine and Piperidine Carbamates as Efficient, Covalent Inhibitors of Monoacylglycerol Lipase. J Med Chem60:9860-9873 (2017) [PubMed] Article Butler, CR; Beck, EM; Harris, A; Huang, Z; McAllister, LA; Am Ende, CW; Fennell, K; Foley, TL; Fonseca, K; Hawrylik, SJ; Johnson, DS; Knafels, JD; Mente, S; Noell, GS; Pandit, J; Phillips, TB; Piro, JR; Rogers, BN; Samad, TA; Wang, J; Wan, S; Brodney, MA Azetidine and Piperidine Carbamates as Efficient, Covalent Inhibitors of Monoacylglycerol Lipase. J Med Chem60:9860-9873 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 |
|---|
| Name: | Fatty-acid amide hydrolase 1 |
|---|
| Synonyms: | Anandamide amidohydrolase | Anandamide amidohydrolase 1 | FAAH | FAAH1 | FAAH1_HUMAN | Fatty Acid Amide Hydrolase (FAAH) | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Oleamide hydrolase 1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 63071.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O00519 |
|---|
| Residue: | 579 |
|---|
| Sequence: | MVQYELWAALPGASGVALACCFVAAAVALRWSGRRTARGAVVRARQRQRAGLENMDRAAQ
RFRLQNPDLDSEALLALPLPQLVQKLHSRELAPEAVLFTYVGKAWEVNKGTNCVTSYLAD
CETQLSQAPRQGLLYGVPVSLKECFTYKGQDSTLGLSLNEGVPAECDSVVVHVLKLQGAV
PFVHTNVPQSMFSYDCSNPLFGQTVNPWKSSKSPGGSSGGEGALIGSGGSPLGLGTDIGG
SIRFPSSFCGICGLKPTGNRLSKSGLKGCVYGQEAVRLSVGPMARDVESLALCLRALLCE
DMFRLDPTVPPLPFREEVYTSSQPLRVGYYETDNYTMPSPAMRRAVLETKQSLEAAGHTL
VPFLPSNIPHALETLSTGGLFSDGGHTFLQNFKGDFVDPCLGDLVSILKLPQWLKGLLAF
LVKPLLPRLSAFLSNMKSRSAGKLWELQHEIEVYRKTVIAQWRALDLDVVLTPMLAPALD
LNAPGRATGAVSYTMLYNCLDFPAGVVPVTTVTAEDEAQMEHYRGYFGDIWDKMLQKGMK
KSVGLPVAVQCVALPWQEELCLRFMREVERLMTPEKQSS
|
|
|
|---|
| BDBM50250859 |
|---|
| n/a |
|---|
| Name | BDBM50250859 |
|---|
| Synonyms: | CHEMBL4097203 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H11F8N3O2 |
|---|
| Mol. Mass. | 429.2647 |
|---|
| SMILES | Fc1ccc(cc1F)-n1ccc(n1)C1CN(C1)C(=O)OC(C(F)(F)F)C(F)(F)F |
|---|
| Structure | 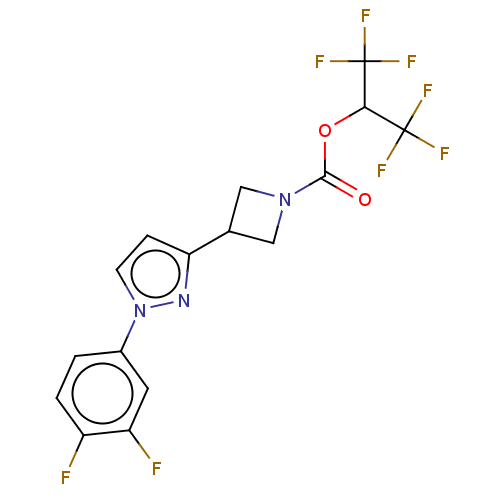
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Butler, CR; Beck, EM; Harris, A; Huang, Z; McAllister, LA; Am Ende, CW; Fennell, K; Foley, TL; Fonseca, K; Hawrylik, SJ; Johnson, DS; Knafels, JD; Mente, S; Noell, GS; Pandit, J; Phillips, TB; Piro, JR; Rogers, BN; Samad, TA; Wang, J; Wan, S; Brodney, MA Azetidine and Piperidine Carbamates as Efficient, Covalent Inhibitors of Monoacylglycerol Lipase. J Med Chem60:9860-9873 (2017) [PubMed] Article
Butler, CR; Beck, EM; Harris, A; Huang, Z; McAllister, LA; Am Ende, CW; Fennell, K; Foley, TL; Fonseca, K; Hawrylik, SJ; Johnson, DS; Knafels, JD; Mente, S; Noell, GS; Pandit, J; Phillips, TB; Piro, JR; Rogers, BN; Samad, TA; Wang, J; Wan, S; Brodney, MA Azetidine and Piperidine Carbamates as Efficient, Covalent Inhibitors of Monoacylglycerol Lipase. J Med Chem60:9860-9873 (2017) [PubMed] Article