| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lactase/phlorizin hydrolase |
|---|
| Ligand | BDBM50031482 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_96426 (CHEMBL706374) |
|---|
| IC50 | <50000±n/a nM |
|---|
| Citation |  Asano, N; Kizu, H; Oseki, K; Tomioka, E; Matsui, K; Okamoto, M; Baba, M N-alkylated nitrogen-in-the-ring sugars: conformational basis of inhibition of glycosidases and HIV-1 replication. J Med Chem38:2349-56 (1995) [PubMed] Asano, N; Kizu, H; Oseki, K; Tomioka, E; Matsui, K; Okamoto, M; Baba, M N-alkylated nitrogen-in-the-ring sugars: conformational basis of inhibition of glycosidases and HIV-1 replication. J Med Chem38:2349-56 (1995) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lactase/phlorizin hydrolase |
|---|
| Name: | Lactase/phlorizin hydrolase |
|---|
| Synonyms: | LPH_RAT | Lactase-glycosylceramidase | Lct | Lph |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 217253.31 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_96426 |
|---|
| Residue: | 1928 |
|---|
| Sequence: | MELPWTALFLSTVLLGLSCQGSDWESDRNFISAAGPLTNDLVLNLNYPPGKQGSDVVSGN
TDHLLCQQPLPSFLSQYFSSLRASQVTHYKVLLSWAQLLPTGSSKNPDQEAVQCYRQLLQ
SLKDAQLEPMVVLCHQTPPTSSAIQREGAFADLFADYATLAFQSFGDLVEIWFTFSDLEK
VIMDLPHKDLKASALQTLSNAHRRAFEIYHRKFSSQGGKLSVVLKAEDIPELLPDPALAA
LVQGSVDFLSLDLSYECQSVATLPQKLSELQNLEPKVKVFIYTLKLEDCPATGTSPSSLL
ISLLEAINKDQIQTVGFDVNAFLSCTSNSEESPSCSLTDSLALQTEQQQETAVPSSPGSA
YQRVWAAFANQSREERDAFLQDVFPEGFLWGISTGAFNVEGGWAEGGRGPSIWDHYGNLN
AAEGQATAKVASDSYHKPASDVALLRGIRAQVYKFSISWSGLFPLGQKSTPNRQGVAYYN
KLIDRLLDSHIEPMATLFHWDLPQALQEQGGWQNESVVEAFLDYAAFCFSTFGDRVKLWV
TFHEPWVMSYAGYGTGQHAPAISDPGMASFKVAHLILKAHARTWHLYDLHHRLQQQGRVG
IVLNSDLAEPLDRKSPQDLAAAERFLHFMLGWFAHPIFVDGDYPTTSAQIQHINQQCGHP
LAQLPEFTEAEKRLLKGSADFLGLSHYTSRLISKAGRQTCTSSYDNIGGFSQHVDPEWPQ
TASPWIRVVPWGIRRLLRFASMEYTKGKLPIFLAGNGMPVGEEADLFDDSVRVNYFNWYI
NEVLKAVKEDLVDVRSYIVRSLIDGYEGPLGFSQRFGLYHVNFNDSSRPRTPRKSAYLFT
SIIEKNGFSAKKVKRNPLPVRADFTSRARVTDSLPSEVPSKAKISVEKFSKQPRFERDLF
YDGRFRDDFLWGVSSSPYQIEGGWNADGKGPSIWDNFTHTPGNGVKDNATGDVACDSYHQ
LDADLNILRTLKVKSYRFSISWSRIFPTGRNSTINKQGVDYYNRLIDSLVDNNIFPMVTL
FHWDLPQALQDIGGWENPSLIELFDSYADYCFKTFGDRVKFWMTFNEPWCHVVLGYSSGI
FPPSVQEPGWLPYKVSHIVIKAHARVYHTYDEKYRSEQKGVISLSLNTHWAEPKDPGLQR
DVEAADRMLQFTMGWFAHPIFKNGDYPDVMKWTVGNRSELQHLASSRLPTFTEEEKNYVR
GTADVFCHNTYTSVFVQHSTPRLNPPSYDDDMELKLIEMNSSTGVMHQDVPWGTRRLLNW
IKEEYGNIPIYITENGQGLENPTLDDTERIFYHKTYINEALKAYKLDGVDLRGYSAWTLM
DDFEWLLGYTMRFGLYYVDFNHVSRPRTARASARYYPDLIANNGMPLAREDEFLYGEFPK
GFIWSAASASYQVEGAWRADGKGLSIWDTFSHTPLRIGNDDNGDVACDSYHKIAEDVVAL
QNLGVSHYRFSIAWSRILPDGTTKFINEAGLSYYVRFIDALLAAGITPQVTIYHWDLPQA
LQDVGGWENETIVQRFKEYADVLFQRLGDRVKFWITLNEPFVIAAQGYGTGVSAPGISFR
PGTAPYIAGHNLIKAHAEAWHLYNDVYRARQGGTISITISSDWGEPRDPTNREHVEAARS
YVQFMGGWFAHPIFKNGDYPEVMKTRIRDRSLGAGLNKSRLPEFTESEKSRIKGTFDFFG
FNHNTTVLAYNLDYPAAFSSFDADRGVASIADSSWPVSGSFWLKVTPFGFRRILNWLKEE
YNNPPIYVTENGVSRRGEPELNDTDRIYYLRSYINEALKAVHDKVDLRGYTVWSIMDNFE
WATGFAERFGVHFVNRSDPSLPRIPRASAKFYATIVRCNGFPDPAQGPHPCLQQPEDAAP
TASPVQSEVPFLGLMLGIAEAQTALYVLFALLLLGACSLAFLTYNTGRRSKQGNAQPSQH
QLSPISSF
|
|
|
|---|
| BDBM50031482 |
|---|
| n/a |
|---|
| Name | BDBM50031482 |
|---|
| Synonyms: | (2R,3R,4R,5R)-1-Butyl-2,5-bis-hydroxymethyl-pyrrolidine-3,4-diol | CHEMBL77918 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H21NO4 |
|---|
| Mol. Mass. | 219.278 |
|---|
| SMILES | CCCCN1[C@H](CO)[C@@H](O)[C@H](O)[C@H]1CO |
|---|
| Structure | 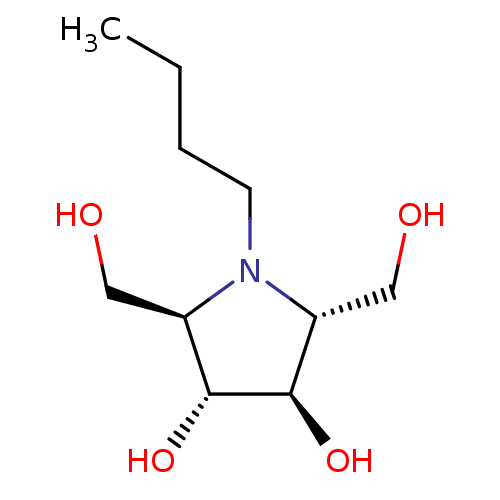
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Asano, N; Kizu, H; Oseki, K; Tomioka, E; Matsui, K; Okamoto, M; Baba, M N-alkylated nitrogen-in-the-ring sugars: conformational basis of inhibition of glycosidases and HIV-1 replication. J Med Chem38:2349-56 (1995) [PubMed]
Asano, N; Kizu, H; Oseki, K; Tomioka, E; Matsui, K; Okamoto, M; Baba, M N-alkylated nitrogen-in-the-ring sugars: conformational basis of inhibition of glycosidases and HIV-1 replication. J Med Chem38:2349-56 (1995) [PubMed]