| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Phospholipase A2 |
|---|
| Ligand | BDBM50053136 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_156184 (CHEMBL760953) |
|---|
| IC50 | 1.4±n/a nM |
|---|
| Citation |  Hagishita, S; Yamada, M; Shirahase, K; Okada, T; Murakami, Y; Ito, Y; Matsuura, T; Wada, M; Kato, T; Ueno, M; Chikazawa, Y; Yamada, K; Ono, T; Teshirogi, I; Ohtani, M Potent inhibitors of secretory phospholipase A2: synthesis and inhibitory activities of indolizine and indene derivatives. J Med Chem39:3636-58 (1996) [PubMed] Article Hagishita, S; Yamada, M; Shirahase, K; Okada, T; Murakami, Y; Ito, Y; Matsuura, T; Wada, M; Kato, T; Ueno, M; Chikazawa, Y; Yamada, K; Ono, T; Teshirogi, I; Ohtani, M Potent inhibitors of secretory phospholipase A2: synthesis and inhibitory activities of indolizine and indene derivatives. J Med Chem39:3636-58 (1996) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Phospholipase A2 |
|---|
| Name: | Phospholipase A2 |
|---|
| Synonyms: | Group IB phospholipase A2 | PA21B_HUMAN | PLA2 | PLA2A | PLA2G1B | PPLA2 | Phosphatidylcholine 2-acylhydrolase 1B | Phospholipase A2 (PLA2) | Phospholipase A2 group 1B | phospholipase A2 precursor |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 16364.13 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P04054 |
|---|
| Residue: | 148 |
|---|
| Sequence: | MKLLVLAVLLTVAAADSGISPRAVWQFRKMIKCVIPGSDPFLEYNNYGCYCGLGGSGTPV
DELDKCCQTHDNCYDQAKKLDSCKFLLDNPYTHTYSYSCSGSAITCSSKNKECEAFICNC
DRNAAICFSKAPYNKAHKNLDTKKYCQS
|
|
|
|---|
| BDBM50053136 |
|---|
| n/a |
|---|
| Name | BDBM50053136 |
|---|
| Synonyms: | (1-Aminooxalyl-3-biphenyl-2-ylmethyl-2-methyl-indolizin-8-yloxy)-acetic acid methyl ester | CHEMBL121627 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H24N2O5 |
|---|
| Mol. Mass. | 456.4899 |
|---|
| SMILES | COC(=O)COc1cccn2c(Cc3ccccc3-c3ccccc3)c(C)c(C(=O)C(N)=O)c12 |
|---|
| Structure | 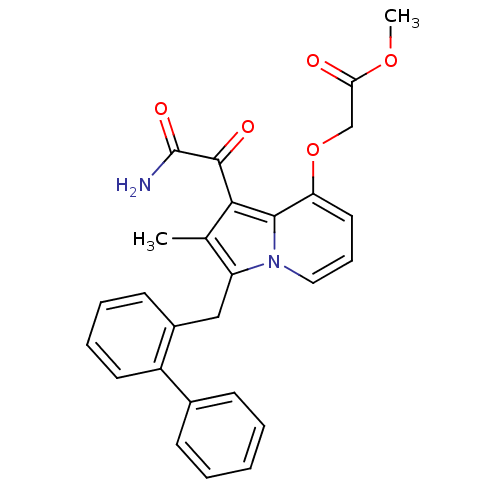
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hagishita, S; Yamada, M; Shirahase, K; Okada, T; Murakami, Y; Ito, Y; Matsuura, T; Wada, M; Kato, T; Ueno, M; Chikazawa, Y; Yamada, K; Ono, T; Teshirogi, I; Ohtani, M Potent inhibitors of secretory phospholipase A2: synthesis and inhibitory activities of indolizine and indene derivatives. J Med Chem39:3636-58 (1996) [PubMed] Article
Hagishita, S; Yamada, M; Shirahase, K; Okada, T; Murakami, Y; Ito, Y; Matsuura, T; Wada, M; Kato, T; Ueno, M; Chikazawa, Y; Yamada, K; Ono, T; Teshirogi, I; Ohtani, M Potent inhibitors of secretory phospholipase A2: synthesis and inhibitory activities of indolizine and indene derivatives. J Med Chem39:3636-58 (1996) [PubMed] Article