| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prostaglandin G/H synthase 2 |
|---|
| Ligand | BDBM50029616 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_159901 (CHEMBL768683) |
|---|
| IC50 | 90±n/a nM |
|---|
| Citation |  Lazer, ES; Miao, CK; Cywin, CL; Sorcek, R; Wong, HC; Meng, Z; Potocki, I; Hoermann, M; Snow, RJ; Tschantz, MA; Kelly, TA; McNeil, DW; Coutts, SJ; Churchill, L; Graham, AG; David, E; Grob, PM; Engel, W; Meier, H; Trummlitz, G Effect of structural modification of enol-carboxamide-type nonsteroidal antiinflammatory drugs on COX-2/COX-1 selectivity. J Med Chem40:980-9 (1997) [PubMed] Article Lazer, ES; Miao, CK; Cywin, CL; Sorcek, R; Wong, HC; Meng, Z; Potocki, I; Hoermann, M; Snow, RJ; Tschantz, MA; Kelly, TA; McNeil, DW; Coutts, SJ; Churchill, L; Graham, AG; David, E; Grob, PM; Engel, W; Meier, H; Trummlitz, G Effect of structural modification of enol-carboxamide-type nonsteroidal antiinflammatory drugs on COX-2/COX-1 selectivity. J Med Chem40:980-9 (1997) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prostaglandin G/H synthase 2 |
|---|
| Name: | Prostaglandin G/H synthase 2 |
|---|
| Synonyms: | COX2 | Cyclooxygenase | Cyclooxygenase 2 (COX-2) | Cyclooxygenase-2 | Cyclooxygenase-2 (COX-2 AA) | Cyclooxygenase-2 (COX-2 AEA) | Cyclooxygenase-2 (COX-2) | PGH synthase 2 | PGH2_HUMAN | PGHS-2 | PHS II | PTGS2 | Prostaglandin E synthase/G/H synthase 2 | Prostaglandin H2 synthase 2 | Prostaglandin-endoperoxide synthase 2 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 69003.89 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Recombinant Cox-2 provided by Cayman (Cayman Chemical Co.,Ann Arbor, MI). |
|---|
| Residue: | 604 |
|---|
| Sequence: | MLARALLLCAVLALSHTANPCCSHPCQNRGVCMSVGFDQYKCDCTRTGFYGENCSTPEFL
TRIKLFLKPTPNTVHYILTHFKGFWNVVNNIPFLRNAIMSYVLTSRSHLIDSPPTYNADY
GYKSWEAFSNLSYYTRALPPVPDDCPTPLGVKGKKQLPDSNEIVEKLLLRRKFIPDPQGS
NMMFAFFAQHFTHQFFKTDHKRGPAFTNGLGHGVDLNHIYGETLARQRKLRLFKDGKMKY
QIIDGEMYPPTVKDTQAEMIYPPQVPEHLRFAVGQEVFGLVPGLMMYATIWLREHNRVCD
VLKQEHPEWGDEQLFQTSRLILIGETIKIVIEDYVQHLSGYHFKLKFDPELLFNKQFQYQ
NRIAAEFNTLYHWHPLLPDTFQIHDQKYNYQQFIYNNSILLEHGITQFVESFTRQIAGRV
AGGRNVPPAVQKVSQASIDQSRQMKYQSFNEYRKRFMLKPYESFEELTGEKEMSAELEAL
YGDIDAVELYPALLVEKPRPDAIFGETMVEVGAPFSLKGLMGNVICSPAYWKPSTFGGEV
GFQIINTASIQSLICNNVKGCPFTSFSVPDPELIKTVTINASSSRSGLDDINPTVLLKER
STEL
|
|
|
|---|
| BDBM50029616 |
|---|
| n/a |
|---|
| Name | BDBM50029616 |
|---|
| Synonyms: | 5-(4-Fluoro-phenyl)-1-(4-methanesulfonyl-phenyl)-3-trifluoromethyl-1H-pyrazole | 5-(4-fluorophenyl)-1-(4-(methylsulfonyl)phenyl)-3-(trifluoromethyl)-1H-pyrazole | CHEMBL274990 | SC-58125 | SC58125 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H12F4N2O2S |
|---|
| Mol. Mass. | 384.348 |
|---|
| SMILES | CS(=O)(=O)c1ccc(cc1)-n1nc(cc1-c1ccc(F)cc1)C(F)(F)F |
|---|
| Structure | 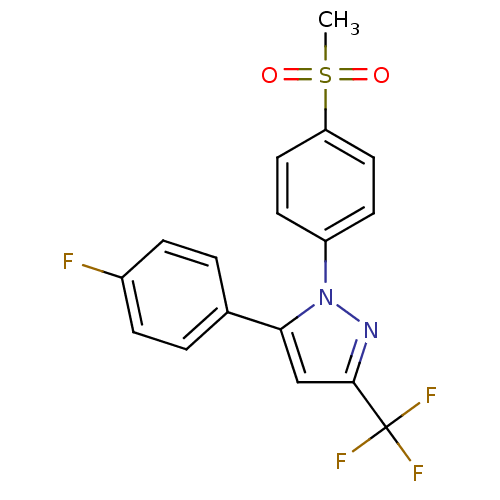
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lazer, ES; Miao, CK; Cywin, CL; Sorcek, R; Wong, HC; Meng, Z; Potocki, I; Hoermann, M; Snow, RJ; Tschantz, MA; Kelly, TA; McNeil, DW; Coutts, SJ; Churchill, L; Graham, AG; David, E; Grob, PM; Engel, W; Meier, H; Trummlitz, G Effect of structural modification of enol-carboxamide-type nonsteroidal antiinflammatory drugs on COX-2/COX-1 selectivity. J Med Chem40:980-9 (1997) [PubMed] Article
Lazer, ES; Miao, CK; Cywin, CL; Sorcek, R; Wong, HC; Meng, Z; Potocki, I; Hoermann, M; Snow, RJ; Tschantz, MA; Kelly, TA; McNeil, DW; Coutts, SJ; Churchill, L; Graham, AG; David, E; Grob, PM; Engel, W; Meier, H; Trummlitz, G Effect of structural modification of enol-carboxamide-type nonsteroidal antiinflammatory drugs on COX-2/COX-1 selectivity. J Med Chem40:980-9 (1997) [PubMed] Article