

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Prothrombin | ||
| Ligand | BDBM50057830 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_208501 (CHEMBL811976) | ||
| Ki | 3000±n/a nM | ||
| Citation |  Tucker, TJ; Lumma, WC; Lewis, SD; Gardell, SJ; Lucas, BJ; Baskin, EP; Woltmann, R; Lynch, JJ; Lyle, EA; Appleby, SD; Chen, IW; Dancheck, KB; Vacca, JP Potent noncovalent thrombin inhibitors that utilize the unique amino acid D-dicyclohexylalanine in the P3 position. Implications on oral bioavailability and antithrombotic efficacy. J Med Chem40:1565-9 (1997) [PubMed] Article Tucker, TJ; Lumma, WC; Lewis, SD; Gardell, SJ; Lucas, BJ; Baskin, EP; Woltmann, R; Lynch, JJ; Lyle, EA; Appleby, SD; Chen, IW; Dancheck, KB; Vacca, JP Potent noncovalent thrombin inhibitors that utilize the unique amino acid D-dicyclohexylalanine in the P3 position. Implications on oral bioavailability and antithrombotic efficacy. J Med Chem40:1565-9 (1997) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Prothrombin | |||
| Name: | Prothrombin | ||
| Synonyms: | Activation peptide fragment 1 | Activation peptide fragment 2 | Coagulation factor II | F2 | Prothrombin precursor | THRB_HUMAN | Thrombin heavy chain | Thrombin light chain | ||
| Type: | Protein | ||
| Mol. Mass.: | 70029.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P00734 | ||
| Residue: | 622 | ||
| Sequence: |
| ||
| BDBM50057830 | |||
| n/a | |||
| Name | BDBM50057830 | ||
| Synonyms: | ((S)-2-{(S)-2-[(4-Amino-cyclohexylmethyl)-carbamoyl]-pyrrolidin-1-yl}-1-dicyclohexylmethyl-2-oxo-ethyl)-carbamic acid tert-butyl ester | CHEMBL289333 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C32H56N4O4 | ||
| Mol. Mass. | 560.8114 | ||
| SMILES | CC(C)(C)OC(=O)N[C@@H](C(C1CCCCC1)C1CCCCC1)C(=O)N1CCC[C@H]1C(=O)NCC1CCC(N)CC1 |wU:8.7,28.31,(4.89,-7.9,;5.69,-6.58,;4.94,-5.24,;6.75,-7.68,;7.68,-6.97,;8.02,-5.27,;7.26,-3.92,;9.56,-5.29,;10.89,-4.53,;10.89,-2.99,;9.56,-2.21,;9.56,-.67,;8.22,.1,;6.9,-.67,;6.9,-2.21,;8.22,-2.98,;12.23,-2.22,;13.55,-2.99,;14.9,-2.22,;14.9,-.68,;13.57,.09,;12.22,-.68,;12.23,-5.3,;12.23,-6.84,;13.71,-4.89,;14.26,-3.44,;15.79,-3.51,;16.21,-5,;14.92,-5.85,;14.85,-7.39,;13.49,-8.1,;16.15,-8.21,;17.52,-7.51,;18.81,-8.32,;17.75,-9.44,;18.84,-10.92,;18.78,-12.17,;19.86,-13.25,;19.86,-11.05,;18.78,-9.66,)| | ||
| Structure | 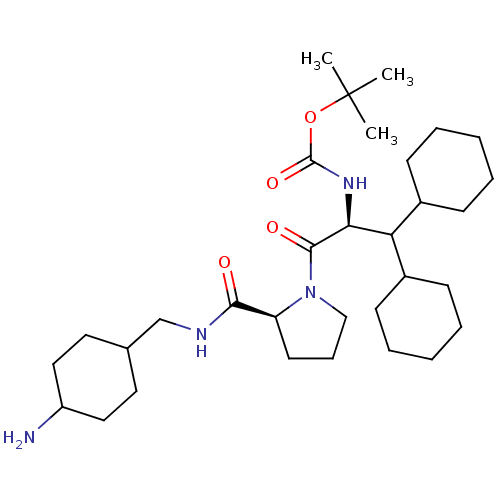
| ||