

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Matrix metalloproteinase-9 | ||
| Ligand | BDBM50073892 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_105232 | ||
| IC50 | 18±n/a nM | ||
| Citation |  Fink, CA; Carlson, JE; Boehm, C; McTaggart, P; Qiao, Y; Doughty, J; Ganu, V; Melton, R; Goldberg, R Design and synthesis of thiol containing inhibitors of matrix metalloproteinases. Bioorg Med Chem Lett9:195-200 (1999) [PubMed] Fink, CA; Carlson, JE; Boehm, C; McTaggart, P; Qiao, Y; Doughty, J; Ganu, V; Melton, R; Goldberg, R Design and synthesis of thiol containing inhibitors of matrix metalloproteinases. Bioorg Med Chem Lett9:195-200 (1999) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Matrix metalloproteinase-9 | |||
| Name: | Matrix metalloproteinase-9 | ||
| Synonyms: | MMP9_RAT | Matrix metalloproteinase 9 | Matrix metalloproteinase-9 (MMP-9) | Mmp9 | ||
| Type: | n/a | ||
| Mol. Mass.: | 78603.01 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | P50282 | ||
| Residue: | 708 | ||
| Sequence: |
| ||
| BDBM50073892 | |||
| n/a | |||
| Name | BDBM50073892 | ||
| Synonyms: | 4-Ethoxy-1-mercaptomethyl-cyclohexanecarboxylic acid ((S)-1-methylcarbamoyl-2-phenyl-ethyl)-amide | CHEMBL61610 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H30N2O3S | ||
| Mol. Mass. | 378.529 | ||
| SMILES | CCO[C@H]1CC[C@](CS)(CC1)C(=O)N[C@@H](Cc1ccccc1)C(=O)NC |wU:6.11,14.14,3.2,wD:6.6,(-5.92,-8.71,;-5.92,-7.17,;-4.61,-6.43,;-4.61,-4.89,;-5.92,-4.12,;-5.92,-2.58,;-4.61,-1.81,;-5.92,-1.04,;-5.92,.5,;-3.26,-2.58,;-3.26,-4.12,;-3.26,-1.04,;-3.26,.5,;-1.91,-1.81,;-.6,-1.04,;-.6,.5,;.17,1.85,;-.63,3.17,;.11,4.51,;1.65,4.54,;2.45,3.23,;1.71,1.88,;.75,-1.81,;.75,-3.35,;2.06,-1.04,;2.06,.5,)| | ||
| Structure | 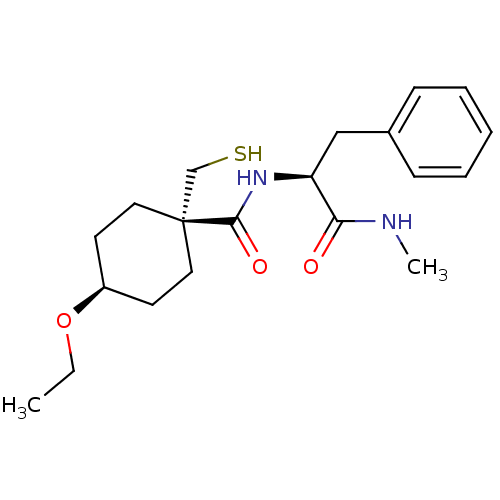
| ||