

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Corticotropin-releasing factor receptor 1 | ||
| Ligand | BDBM50074492 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_51114 (CHEMBL664960) | ||
| Ki | 0.9±n/a nM | ||
| Citation |  Chorvat, RJ; Bakthavatchalam, R; Beck, JP; Gilligan, PJ; Wilde, RG; Cocuzza, AJ; Hobbs, FW; Cheeseman, RS; Curry, M; Rescinito, JP; Krenitsky, P; Chidester, D; Yarem, JA; Klaczkiewicz, JD; Hodge, CN; Aldrich, PE; Wasserman, ZR; Fernandez, CH; Zaczek, R; Fitzgerald, LW; Huang, SM; Shen, HL; Wong, YN; Chien, BM; Arvanitis, A Synthesis, corticotropin-releasing factor receptor binding affinity, and pharmacokinetic properties of triazolo-, imidazo-, and pyrrolopyrimidines and -pyridines. J Med Chem42:833-48 (1999) [PubMed] Article Chorvat, RJ; Bakthavatchalam, R; Beck, JP; Gilligan, PJ; Wilde, RG; Cocuzza, AJ; Hobbs, FW; Cheeseman, RS; Curry, M; Rescinito, JP; Krenitsky, P; Chidester, D; Yarem, JA; Klaczkiewicz, JD; Hodge, CN; Aldrich, PE; Wasserman, ZR; Fernandez, CH; Zaczek, R; Fitzgerald, LW; Huang, SM; Shen, HL; Wong, YN; Chien, BM; Arvanitis, A Synthesis, corticotropin-releasing factor receptor binding affinity, and pharmacokinetic properties of triazolo-, imidazo-, and pyrrolopyrimidines and -pyridines. J Med Chem42:833-48 (1999) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Corticotropin-releasing factor receptor 1 | |||
| Name: | Corticotropin-releasing factor receptor 1 | ||
| Synonyms: | CRF-R | CRF-R2 Alpha | CRF1 | CRFR | CRFR1 | CRFR1_HUMAN | CRH-R 1 | CRHR | CRHR1 | Corticotropin releasing factor receptor 1 | Corticotropin-releasing factor receptor 1 (CRF-1) | Corticotropin-releasing factor receptor 1 (CRF1) | Corticotropin-releasing hormone receptor 1 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 50744.31 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P34998 | ||
| Residue: | 444 | ||
| Sequence: |
| ||
| BDBM50074492 | |||
| n/a | |||
| Name | BDBM50074492 | ||
| Synonyms: | CHEMBL168554 | [9-(2-Bromo-4-isopropyl-phenyl)-2,8-dimethyl-9H-purin-6-yl]-butyl-ethyl-amine | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H30BrN5 | ||
| Mol. Mass. | 444.411 | ||
| SMILES | CCCCN(CC)c1nc(C)nc2n(c(C)nc12)-c1ccc(cc1Br)C(C)C |(12.64,1.78,;12.64,.23,;11.31,-.54,;11.31,-2.08,;9.99,-2.85,;8.64,-2.08,;7.32,-2.85,;9.99,-4.38,;8.64,-5.15,;8.64,-6.69,;7.31,-7.46,;9.99,-7.46,;11.31,-6.69,;12.76,-7.15,;13.67,-5.92,;15.21,-5.94,;12.76,-4.68,;11.31,-5.15,;13.39,-8.57,;12.49,-9.81,;13.11,-11.22,;14.65,-11.38,;15.56,-10.13,;14.93,-8.73,;15.84,-7.48,;15.27,-12.8,;16.79,-12.97,;14.35,-14.04,)| | ||
| Structure | 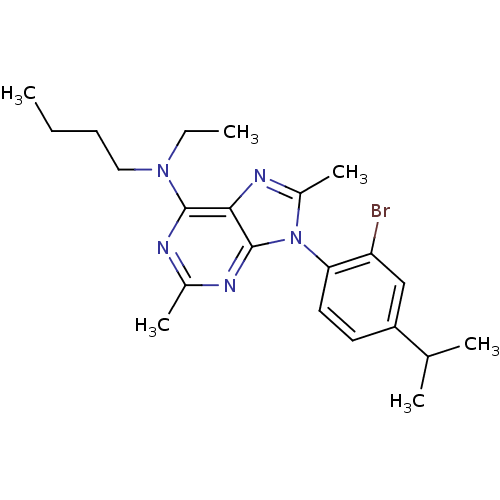
| ||