| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prothrombin |
|---|
| Ligand | BDBM50077058 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_208529 |
|---|
| Ki | 26±n/a nM |
|---|
| Citation |  Ambler, J; Brown, L; Cockcroft, XL; Grütter, M; Hayler, J; Janus, D; Jones, D; Kane, P; Menear, K; Priestle, J; Smith, G; Talbot, M; Walker, CV; Wathey, B Optimisation of the P2 pharmacophore in a series of thrombin inhibitors: ion-dipole interactions with lysine 60G. Bioorg Med Chem Lett9:1317-22 (1999) [PubMed] Ambler, J; Brown, L; Cockcroft, XL; Grütter, M; Hayler, J; Janus, D; Jones, D; Kane, P; Menear, K; Priestle, J; Smith, G; Talbot, M; Walker, CV; Wathey, B Optimisation of the P2 pharmacophore in a series of thrombin inhibitors: ion-dipole interactions with lysine 60G. Bioorg Med Chem Lett9:1317-22 (1999) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prothrombin |
|---|
| Name: | Prothrombin |
|---|
| Synonyms: | Activation peptide fragment 1 | Activation peptide fragment 2 | Coagulation factor II | F2 | Prothrombin precursor | THRB_HUMAN | Thrombin heavy chain | Thrombin light chain |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 70029.57 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P00734 |
|---|
| Residue: | 622 |
|---|
| Sequence: | MAHVRGLQLPGCLALAALCSLVHSQHVFLAPQQARSLLQRVRRANTFLEEVRKGNLEREC
VEETCSYEEAFEALESSTATDVFWAKYTACETARTPRDKLAACLEGNCAEGLGTNYRGHV
NITRSGIECQLWRSRYPHKPEINSTTHPGADLQENFCRNPDSSTTGPWCYTTDPTVRRQE
CSIPVCGQDQVTVAMTPRSEGSSVNLSPPLEQCVPDRGQQYQGRLAVTTHGLPCLAWASA
QAKALSKHQDFNSAVQLVENFCRNPDGDEEGVWCYVAGKPGDFGYCDLNYCEEAVEEETG
DGLDEDSDRAIEGRTATSEYQTFFNPRTFGSGEADCGLRPLFEKKSLEDKTERELLESYI
DGRIVEGSDAEIGMSPWQVMLFRKSPQELLCGASLISDRWVLTAAHCLLYPPWDKNFTEN
DLLVRIGKHSRTRYERNIEKISMLEKIYIHPRYNWRENLDRDIALMKLKKPVAFSDYIHP
VCLPDRETAASLLQAGYKGRVTGWGNLKETWTANVGKGQPSVLQVVNLPIVERPVCKDST
RIRITDNMFCAGYKPDEGKRGDACEGDSGGPFVMKSPFNNRWYQMGIVSWGEGCDRDGKY
GFYTHVFRLKKWIQKVIDQFGE
|
|
|
|---|
| BDBM50077058 |
|---|
| n/a |
|---|
| Name | BDBM50077058 |
|---|
| Synonyms: | 3,3-Dimethyl-1,2,3,4-tetrahydro-quinoline-8-sulfonic acid {(S)-1-benzothiazol-2-ylmethyl-2-[4-(2-fluoro-ethyl)-piperidin-1-yl]-2-oxo-ethyl}-amide | CHEMBL286064 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H35FN4O3S2 |
|---|
| Mol. Mass. | 558.731 |
|---|
| SMILES | CC1(C)CNc2c(C1)cccc2S(=O)(=O)N[C@@H](Cc1nc2ccccc2s1)C(=O)N1CCC(CCF)CC1 |
|---|
| Structure | 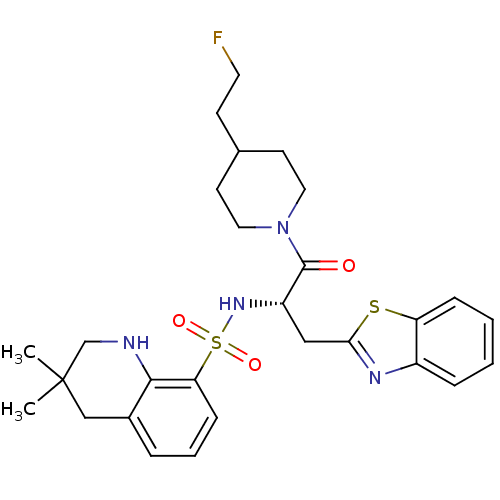
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ambler, J; Brown, L; Cockcroft, XL; Grütter, M; Hayler, J; Janus, D; Jones, D; Kane, P; Menear, K; Priestle, J; Smith, G; Talbot, M; Walker, CV; Wathey, B Optimisation of the P2 pharmacophore in a series of thrombin inhibitors: ion-dipole interactions with lysine 60G. Bioorg Med Chem Lett9:1317-22 (1999) [PubMed]
Ambler, J; Brown, L; Cockcroft, XL; Grütter, M; Hayler, J; Janus, D; Jones, D; Kane, P; Menear, K; Priestle, J; Smith, G; Talbot, M; Walker, CV; Wathey, B Optimisation of the P2 pharmacophore in a series of thrombin inhibitors: ion-dipole interactions with lysine 60G. Bioorg Med Chem Lett9:1317-22 (1999) [PubMed]