| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Voltage-dependent L-type calcium channel subunit alpha-1C |
|---|
| Ligand | BDBM50079464 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_214942 (CHEMBL819393) |
|---|
| Ki | 11±n/a nM |
|---|
| Citation |  Natale, NR; Rogers, ME; Staples, R; Triggle, DJ; Rutledge, A Lipophilic 4-isoxazolyl-1,4-dihydropyridines: synthesis and structure-activity relationships. J Med Chem42:3087-93 (1999) [PubMed] Article Natale, NR; Rogers, ME; Staples, R; Triggle, DJ; Rutledge, A Lipophilic 4-isoxazolyl-1,4-dihydropyridines: synthesis and structure-activity relationships. J Med Chem42:3087-93 (1999) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Voltage-dependent L-type calcium channel subunit alpha-1C |
|---|
| Name: | Voltage-dependent L-type calcium channel subunit alpha-1C |
|---|
| Synonyms: | CAC1C_RAT | Cach2 | Cacn2 | Cacna1c | Cacnl1a1 | Calcium channel | Calcium channel (Type L) | Calcium channel diltiazem | Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle | Cchl1a1 | RBC | Rat brain class C | Voltage-dependent L-type calcium channel subunit alpha-1C | Voltage-gated L-type calcium channel | Voltage-gated L-type calcium channel alpha-1C subunit | Voltage-gated calcium channel subunit alpha Cav1.2 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 243492.26 |
|---|
| Organism: | RAT |
|---|
| Description: | Calcium channel 0 RAT::P22002 |
|---|
| Residue: | 2169 |
|---|
| Sequence: | MIRAFAQPSTPPYQPLSSCLSEDTERKFKGKVVHEAQLNCFYISPGGSNYGSPRPAHANM
NANAAAGLAPEHIPTPGAALSWLAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK
KQGGTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPED
DSNATNSNLERVEYLFLIIFTVEAFLKVIAYGLLFHPNAYLRNGWNLLDFIIVVVGLFSA
ILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH
IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIIDVPAEEDPSPCALETGHGRQCQNGT
VCKPGWDGPKHGITNFDNFAFAMLTVFQCITMEGWTDVLYWMQDAMGYELPWVYFVSLVI
FGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE
NEDEGMDEDKPRNMSMPTSETESVNTENVAGGDIEGENCGARLAHRISKSKFSRYWRRWN
RFCRRKCRAAVKSNVFYWLVIFLVFLNTLTIASEHYNQPHWLTEVQDTANKALLALFTAE
MLLKMYSLGLQAYFVSLFNRFDCFIVCGGILETILVETKIMSPLGISCWRCVRLLRIFKI
TRYWNSLSNLVASLLNSLRSIASLLLLLFLFIIIFSLLGMQLFGGKFNFDEMQTRRSTFD
NFPQSLLTVFQILTGEDWNSVMYDGIMAYGGPSFPGMLVCIYFIILFISPNYILLNLFLA
IAVDNLADAESLTSAQKEEEEEKERKKLARTASPEKKQEVMEKPAVEESKEEKIELKSIT
ADGESPPTTKINMDDLQPSENEDKSPHSNPDTAGEEDEEEPEMPVGPRPRPLSELHLKEK
AVPMPEASAFFIFSPNNRFRLQCHRIVNDTIFTNLILFFILLSSISLAAEDPVQHTSFRN
HILFYFDIVFTTIFTIEIALKMTAYGAFLHKGSFCRNYFNILDLLVVSVSLISFGIQSSA
INVVKILRVLRVLRPLRINRAKGLKHVVQCVFVAIRTIGNIVIVTTLLQFMFACIGVQLF
KGKLYTCSDSSKQTEAESKGNYITYKTGEVDHPIIQPRSWENSKFDFDNVLAAMMALFTV
STFEGWPELLYRSIDSHTEDKGPIYNYRVEISIFFIIYIIIIAFFMMNIFVGFVIVTFQE
QGEQEYKNCELDKNQRQCVEYALKARPLPRYIPKNQHQYKVWYVVNSTYFEYLMFVLILL
NTICLAMQHYGQSCLFKIAMNILNMLFTGLFTVEMILKLIAFKPKHYFCDAWNTFDALIV
VGSIVDIAITEVHPAEHTQCSPSMSAEENSRISITFFRLFRVMRLVKLLSRGEGIRTLLW
TFIKSFQALPYVALLIVMLFFIYAVIGMQVFGKIALNDTTEINRNNNFQTFPQAVLLLFR
CATGEAWQDIMLACMPGKKCAPESEPSNSTEGETPCGSSFAVFYFISFYMLCAFLIINLF
VAVIMDNFDYLTRDWSILGPHHLDEFKRIWAEYDPEAKGRIKHLDVVTLLRRIQPPLGFG
KLCPHRVACKRLVSMNMPLNSDGTVMFNATLFALVRTALRIKTEGNLEQANEELRAIIKK
IWKRTSMKLLDQVVPPAGDDEVTVGKFYATFLIQEYFRKFKKRKEQGLVGKPSQRNALSL
QAGLRTLHDIGPEIRRAISGDLTAEEELDKAMKEAVSAASEDDIFRRAGGLFGNHVSYYQ
SDSRSNFPQTFATQRPLHINKTGNNQADTESPSHEKLVDSTFTPSSYSSTGSNANINNAN
NTALGRFPHPAGYSSTVSTVEGHGPPLSPAVRVQEAAWKLSSKRCHSRESQGATVSQDMF
PDETRSSVRLSEEVEYCSEPSLLSTDILSYQDDENRQLTCLEEDKREIQPCPKRSFLRSA
SLGRRASFHLECLKRQKDQGGDISQKTALPLHLVHHQALAVAGLSPLLQRSHSPSTFPRP
RPTPPVTPGSRGRPLQPIPTLRLEGAESSEKLNSSFPSIHCSSWSEETTACSGGSSMARR
ARPVSLTVPSQAGAPGRQFHGSASSLVEAVLISEGLGQFAQDPKFIEVTTQELADACDMT
IEEMENAADNILSGGAQQSPNGTLLPFVNCRDPGQDRAVVPEDESCVYALGRGRSEEALP
DSRSYVSNL
|
|
|
|---|
| BDBM50079464 |
|---|
| n/a |
|---|
| Name | BDBM50079464 |
|---|
| Synonyms: | 2,6-Dimethyl-4-[3-methyl-5-(2-naphthalen-2-yl-ethyl)-isoxazol-4-yl]-1,4-dihydro-pyridine-3,5-dicarboxylic acid diethyl ester | CHEMBL317651 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C29H32N2O5 |
|---|
| Mol. Mass. | 488.5748 |
|---|
| SMILES | CCOC(=O)C1C(c2c(C)noc2CCc2ccc3ccccc3c2)C(C(=O)OCC)=C(C)N=C1C |c:36,t:33| |
|---|
| Structure | 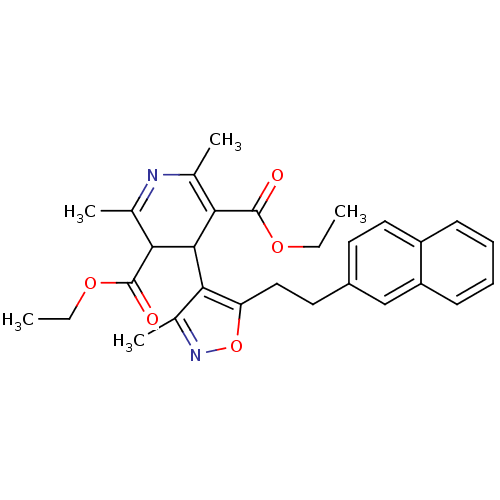
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Natale, NR; Rogers, ME; Staples, R; Triggle, DJ; Rutledge, A Lipophilic 4-isoxazolyl-1,4-dihydropyridines: synthesis and structure-activity relationships. J Med Chem42:3087-93 (1999) [PubMed] Article
Natale, NR; Rogers, ME; Staples, R; Triggle, DJ; Rutledge, A Lipophilic 4-isoxazolyl-1,4-dihydropyridines: synthesis and structure-activity relationships. J Med Chem42:3087-93 (1999) [PubMed] Article