

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Prothrombin | ||
| Ligand | BDBM50084082 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_208331 | ||
| Ki | 115±n/a nM | ||
| Citation |  Lu, T; Soll, RM; Illig, CR; Bone, R; Murphy, L; Spurlino, J; Salemme, FR; Tomczuk, BE Structure-activity and crystallographic analysis of a new class of non-amide-based thrombin inhibitor. Bioorg Med Chem Lett10:79-82 (2000) [PubMed] Lu, T; Soll, RM; Illig, CR; Bone, R; Murphy, L; Spurlino, J; Salemme, FR; Tomczuk, BE Structure-activity and crystallographic analysis of a new class of non-amide-based thrombin inhibitor. Bioorg Med Chem Lett10:79-82 (2000) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Prothrombin | |||
| Name: | Prothrombin | ||
| Synonyms: | Activation peptide fragment 1 | Activation peptide fragment 2 | Coagulation factor II | F2 | Prothrombin precursor | THRB_HUMAN | Thrombin heavy chain | Thrombin light chain | ||
| Type: | Protein | ||
| Mol. Mass.: | 70029.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P00734 | ||
| Residue: | 622 | ||
| Sequence: |
| ||
| BDBM50084082 | |||
| n/a | |||
| Name | BDBM50084082 | ||
| Synonyms: | 3-Nitro-benzenesulfonic acid 3-(1-carbamimidoyl-piperidin-4-ylmethoxy)-5-methyl-phenyl ester | CHEMBL167884 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H24N4O6S | ||
| Mol. Mass. | 448.493 | ||
| SMILES | Cc1cc(OCC2CC[N+](CC2)=C(N)[NH-])cc(OS(=O)(=O)c2cccc(c2)[N+]([O-])=O)c1 |(4.5,-3.85,;4.95,-5.32,;6.46,-5.67,;6.9,-7.14,;8.4,-7.49,;9.45,-6.37,;10.94,-6.72,;11.39,-8.21,;12.88,-8.56,;13.93,-7.45,;13.5,-5.97,;11.99,-5.6,;15.44,-7.8,;16.49,-6.68,;15.86,-9.28,;5.84,-8.26,;4.34,-7.9,;3.3,-9.02,;1.8,-8.67,;2.56,-7.33,;.47,-7.9,;.75,-9.79,;-.76,-9.44,;-1.81,-10.56,;-1.37,-12.04,;.12,-12.39,;1.19,-11.27,;.57,-13.87,;-.48,-14.99,;2.08,-14.23,;3.9,-6.44,)| | ||
| Structure | 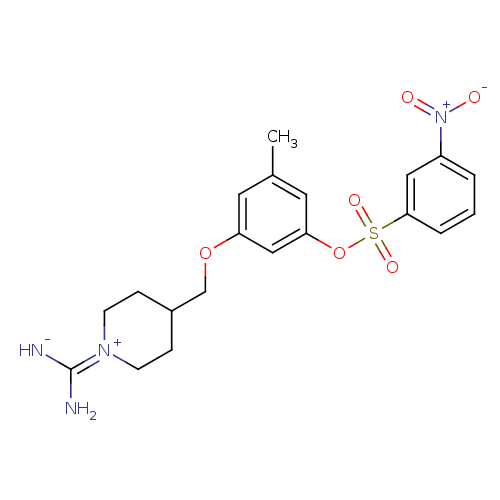
| ||