

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Neuropeptide Y receptor type 5 | ||
| Ligand | BDBM50089038 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_144134 | ||
| IC50 | 1.4±n/a nM | ||
| Citation |  Rueeger, H; Rigollier, P; Yamaguchi, Y; Schmidlin, T; Schilling, W; Criscione, L; Whitebread, S; Chiesi, M; Walker, MW; Dhanoa, D; Islam, I; Zhang, J; Gluchowski, C Design, synthesis and SAR of a series of 2-substituted 4-amino-quinazoline neuropeptide Y Y5 receptor antagonists. Bioorg Med Chem Lett10:1175-9 (2000) [PubMed] Rueeger, H; Rigollier, P; Yamaguchi, Y; Schmidlin, T; Schilling, W; Criscione, L; Whitebread, S; Chiesi, M; Walker, MW; Dhanoa, D; Islam, I; Zhang, J; Gluchowski, C Design, synthesis and SAR of a series of 2-substituted 4-amino-quinazoline neuropeptide Y Y5 receptor antagonists. Bioorg Med Chem Lett10:1175-9 (2000) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Neuropeptide Y receptor type 5 | |||
| Name: | Neuropeptide Y receptor type 5 | ||
| Synonyms: | NPY-Y5 | NPY5R_RAT | Neuropeptide Y receptor type 5 | Npy5r | Npyr5 | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 50435.58 | ||
| Organism: | Rat 6B | ||
| Description: | NPY-Y5 NPY5R Rat 6B::Q63634 | ||
| Residue: | 445 | ||
| Sequence: |
| ||
| BDBM50089038 | |||
| n/a | |||
| Name | BDBM50089038 | ||
| Synonyms: | CGP 71683 | CGP-71683A | CHEMBL17645 | N-{[(1r,4r)-4-{[(4-aminoquinazolin-2-yl)amino]methyl}cyclohexyl]methyl}naphthalene-1-sulfonamide (Compound 1) | Naphthalene-1-sulfonic acid {4-[(4-amino-quinazolin-2-ylamino)-methyl]-cyclohexylmethyl}-amide | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H29N5O2S | ||
| Mol. Mass. | 475.606 | ||
| SMILES | Nc1nc(NC[C@H]2CC[C@H](CNS(=O)(=O)c3cccc4ccccc34)CC2)nc2ccccc12 |wU:6.5,wD:9.9,(2.92,.42,;2.94,-1.13,;4.28,-1.89,;4.29,-3.44,;5.63,-4.22,;6.96,-3.45,;8.29,-4.21,;9.63,-3.44,;10.96,-4.22,;10.95,-5.76,;12.28,-6.54,;13.77,-6.13,;14.86,-7.21,;13.98,-8.47,;15.7,-5.91,;16.12,-8.05,;17.24,-7,;18.73,-7.45,;19.09,-8.94,;17.97,-9.99,;18.33,-11.48,;17.22,-12.56,;15.73,-12.11,;15.38,-10.62,;16.49,-9.56,;9.62,-6.51,;8.29,-5.75,;2.95,-4.23,;1.61,-3.45,;.26,-4.23,;-1.07,-3.46,;-1.07,-1.9,;.26,-1.13,;1.61,-1.9,)| | ||
| Structure | 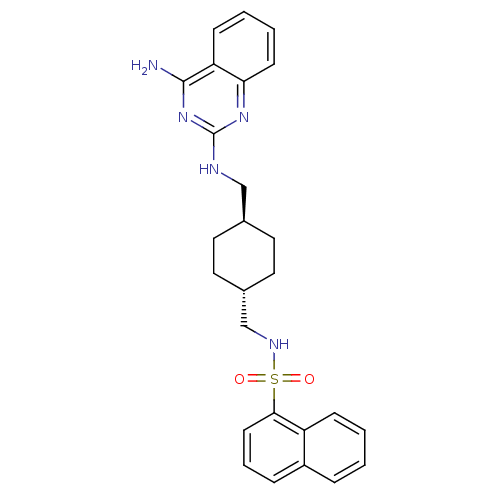
| ||