| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Trypanothione reductase |
|---|
| Ligand | BDBM50015214 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_211812 |
|---|
| IC50 | 133000±n/a nM |
|---|
| Citation |  Chibale, K; Haupt, H; Kendrick, H; Yardley, V; Saravanamuthu, A; Fairlamb, AH; Croft, SL Antiprotozoal and cytotoxicity evaluation of sulfonamide and urea analogues of quinacrine. Bioorg Med Chem Lett11:2655-7 (2001) [PubMed] Chibale, K; Haupt, H; Kendrick, H; Yardley, V; Saravanamuthu, A; Fairlamb, AH; Croft, SL Antiprotozoal and cytotoxicity evaluation of sulfonamide and urea analogues of quinacrine. Bioorg Med Chem Lett11:2655-7 (2001) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Trypanothione reductase |
|---|
| Name: | Trypanothione reductase |
|---|
| Synonyms: | N(1),N(8)-bis(glutathionyl)spermidine reductase | TPR | TR | TYTR_TRYCR |
|---|
| Type: | Homodimer; oxidoreductase |
|---|
| Mol. Mass.: | 53868.26 |
|---|
| Organism: | Trypanosoma cruzi |
|---|
| Description: | n/a |
|---|
| Residue: | 492 |
|---|
| Sequence: | MMSKIFDLVVIGAGSGGLEAAWNAATLYKKRVAVIDVQMVHGPPFFSALGGTCVNVGCVP
KKLMVTGAQYMEHLRESAGFGWEFDRTTLRAEWKKLIAVKDEAVLNINKSYEEMFRDTEG
LEFFLGWGSLESKNVVNVRESADPASAVKERLETENILLASGSWPHMPNIPGIEHCISSN
EAFYLPEPPRRVLTVGGGFISVEFAGIFNAYKPKDGQVTLCYRGEMILRGFDHTLREELT
KQLTANGIQILTKENPAKVELNADGSKSVTFESGKKMDFDLVMMAIGRSPRTKDLQLQNA
GVMIKNGGVQVDEYSRTNVSNIYAIGDVTNRVMLTPVAINEAAALVDTVFGTNPRKTDHT
RVASAVFSIPPIGTCGLIEEVASKRYEVVAVYLSSFTPLMHNISGSKYKTFVAKIITNHS
DGTVLGVHLLGDNAPEIIQGVGICLKLNAKISDFYNTIGVHPTSAEELCSMRTPSYYYVK
GEKMEKPSEASL
|
|
|
|---|
| BDBM50015214 |
|---|
| n/a |
|---|
| Name | BDBM50015214 |
|---|
| Synonyms: | 6-chloro-2-methoxy-9-acridinyl(4-diethylamino-1-methylbutyl)amine | 6-chloro-N-(5-(diethylamino)pentan-2-yl)-2-methoxyacridin-9-amine | CHEMBL7568 | N'-(6-chloro-2-methoxy-acridin-9-yl)-N,N-diethyl-pentane-1,4-diamine | N*4*-(6-Chloro-2-methoxy-acridin-9-yl)-N*1*,N*1*-diethyl-pentane-1,4-diamine | N*4*-(6-Chloro-2-methoxy-acridin-9-yl)-N*1*,N*1*-diethyl-pentane-1,4-diamine (Mepacrine) | N*4*-(6-Chloro-2-methoxy-acridin-9-yl)-N*1*,N*1*-diethyl-pentane-1,4-diamine ; (mepacrine) | N-(4-Hydroxy-phenyl)-acetamide | cid_237 | quinacrine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H30ClN3O |
|---|
| Mol. Mass. | 399.957 |
|---|
| SMILES | CCN(CC)CCCC(C)Nc1c2ccc(Cl)cc2nc2ccc(OC)cc12 |
|---|
| Structure | 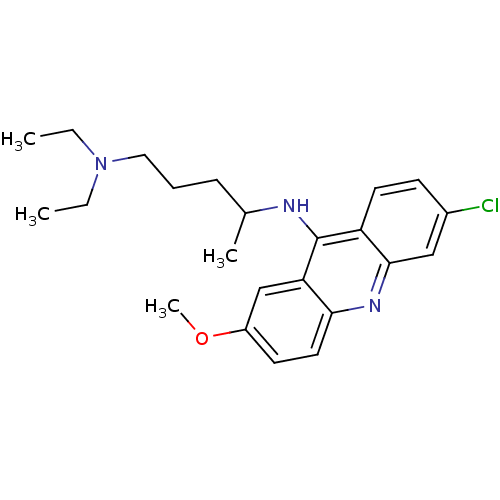
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Chibale, K; Haupt, H; Kendrick, H; Yardley, V; Saravanamuthu, A; Fairlamb, AH; Croft, SL Antiprotozoal and cytotoxicity evaluation of sulfonamide and urea analogues of quinacrine. Bioorg Med Chem Lett11:2655-7 (2001) [PubMed]
Chibale, K; Haupt, H; Kendrick, H; Yardley, V; Saravanamuthu, A; Fairlamb, AH; Croft, SL Antiprotozoal and cytotoxicity evaluation of sulfonamide and urea analogues of quinacrine. Bioorg Med Chem Lett11:2655-7 (2001) [PubMed]