

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Coagulation factor X | ||
| Ligand | BDBM50112506 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_49302 | ||
| Ki | 0.070000±n/a nM | ||
| Citation |  Wu, S; Guilford, WJ; Chou, YL; Griedel, BD; Liang, A; Sakata, S; Shaw, KJ; Trinh, L; Xu, W; Zhao, Z; Morrissey, MM Design and synthesis of aminophenol-based factor Xa inhibitors. Bioorg Med Chem Lett12:1307-10 (2002) [PubMed] Wu, S; Guilford, WJ; Chou, YL; Griedel, BD; Liang, A; Sakata, S; Shaw, KJ; Trinh, L; Xu, W; Zhao, Z; Morrissey, MM Design and synthesis of aminophenol-based factor Xa inhibitors. Bioorg Med Chem Lett12:1307-10 (2002) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Coagulation factor X | |||
| Name: | Coagulation factor X | ||
| Synonyms: | Activated coagulation factor X (FXa) | Activated factor Xa heavy chain | Coagulation factor X precursor | Coagulation factor Xa | F10 | FA10_HUMAN | Factor X heavy chain | Factor X light chain | Factor Xa | Stuart factor | Stuart-Prower factor | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 54726.60 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 488 | ||
| Sequence: |
| ||
| BDBM50112506 | |||
| n/a | |||
| Name | BDBM50112506 | ||
| Synonyms: | 3-(3-{4-[1-(1-Imino-ethyl)-piperidin-4-yloxy]-3-nitro-phenylamino}-2-methyl-propenyl)-benzamidine | CHEMBL27228 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C24H30N6O3 | ||
| Mol. Mass. | 450.5334 | ||
| SMILES | C\C(CNc1ccc(OC2CC[N+](CC2)=C(C)[NH-])c(c1)[N+]([O-])=O)=C/c1cccc(c1)C(N)=N |(8.92,-4.79,;7.58,-5.56,;6.23,-4.79,;6.23,-3.25,;4.91,-2.48,;3.57,-3.25,;2.24,-2.48,;2.24,-.94,;.91,-.17,;-.44,-.94,;-1.76,-.16,;-3.09,-.92,;-3.1,-2.46,;-1.76,-3.24,;-.44,-2.47,;-4.43,-3.23,;-5.78,-2.46,;-4.45,-4.77,;3.57,-.17,;4.91,-.94,;3.55,1.37,;4.9,2.14,;2.22,2.14,;7.56,-7.1,;8.89,-7.87,;8.91,-9.41,;10.25,-10.18,;11.58,-9.4,;11.57,-7.84,;10.22,-7.09,;12.9,-7.07,;12.88,-5.53,;14.23,-7.82,)| | ||
| Structure | 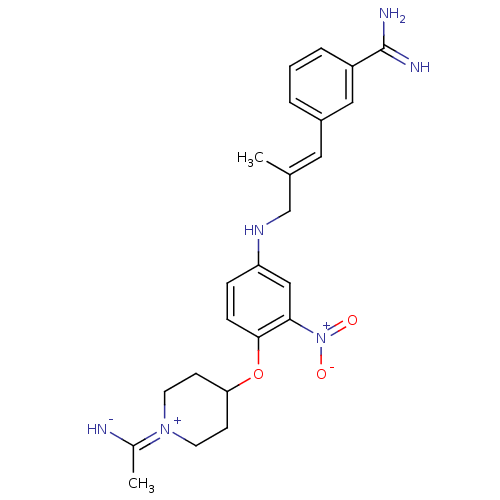
| ||