

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Mitogen-activated protein kinase 11 | ||
| Ligand | BDBM50122910 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_124648 (CHEMBL731909) | ||
| IC50 | 153±n/a nM | ||
| Citation |  Colletti, SL; Frie, JL; Dixon, EC; Singh, SB; Choi, BK; Scapin, G; Fitzgerald, CE; Kumar, S; Nichols, EA; O'Keefe, SJ; O'Neill, EA; Porter, G; Samuel, K; Schmatz, DM; Schwartz, CD; Shoop, WL; Thompson, CM; Thompson, JE; Wang, R; Woods, A; Zaller, DM; Doherty, JB Hybrid-designed inhibitors of p38 MAP kinase utilizing N-arylpyridazinones. J Med Chem46:349-52 (2003) [PubMed] Article Colletti, SL; Frie, JL; Dixon, EC; Singh, SB; Choi, BK; Scapin, G; Fitzgerald, CE; Kumar, S; Nichols, EA; O'Keefe, SJ; O'Neill, EA; Porter, G; Samuel, K; Schmatz, DM; Schwartz, CD; Shoop, WL; Thompson, CM; Thompson, JE; Wang, R; Woods, A; Zaller, DM; Doherty, JB Hybrid-designed inhibitors of p38 MAP kinase utilizing N-arylpyridazinones. J Med Chem46:349-52 (2003) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Mitogen-activated protein kinase 11 | |||
| Name: | Mitogen-activated protein kinase 11 | ||
| Synonyms: | MAP Kinase p38 beta | MK11_MOUSE | Mapk11 | Mitogen-activated protein kinase 11 | Mitogen-activated protein kinase p38 beta | Prkm11 | p38B | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 41390.66 | ||
| Organism: | Mus musculus (mouse) | ||
| Description: | Q9WUI1 | ||
| Residue: | 364 | ||
| Sequence: |
| ||
| BDBM50122910 | |||
| n/a | |||
| Name | BDBM50122910 | ||
| Synonyms: | 2-(2,6-Dichloro-phenyl)-6-[2-(4-fluoro-phenyl)-imidazo[1,2-a]pyridin-3-yl]-2H-pyridazin-3-one | 2-(2,6-dichlorophenyl)-6-(2-(4-fluorophenyl)imidazo[1,2-a]pyridin-3-yl)pyridazin-3(2H)-one | CHEMBL117109 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H13Cl2FN4O | ||
| Mol. Mass. | 451.28 | ||
| SMILES | Fc1ccc(cc1)-c1nc2ccccn2c1-c1ccc(=O)n(n1)-c1c(Cl)cccc1Cl |(-4.28,-10.03,;-3.48,-8.71,;-4.22,-7.36,;-3.43,-6.05,;-1.89,-6.08,;-1.14,-7.42,;-1.94,-8.74,;-1.1,-4.75,;.4,-5.08,;1.17,-3.75,;2.67,-3.44,;3.16,-1.98,;2.13,-.83,;.62,-1.14,;.15,-2.6,;-1.27,-3.22,;-2.6,-2.44,;-3.93,-3.21,;-5.26,-2.43,;-5.26,-.9,;-6.59,-.13,;-3.92,-.13,;-2.59,-.9,;-3.92,1.41,;-5.25,2.18,;-6.59,1.41,;-5.25,3.72,;-3.92,4.49,;-2.59,3.73,;-2.58,2.18,;-1.25,1.41,)| | ||
| Structure | 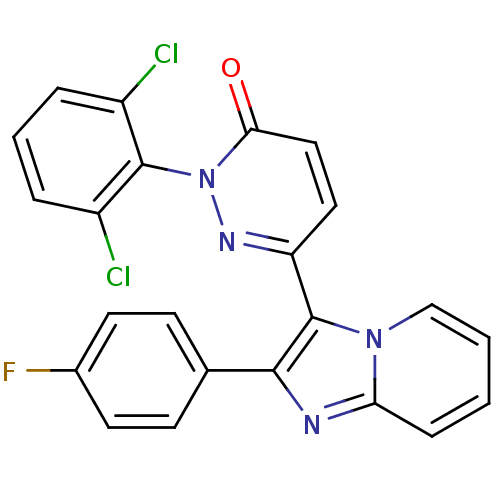
| ||