| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine protease 1 |
|---|
| Ligand | BDBM50123503 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_212685 (CHEMBL815470) |
|---|
| Ki | 118000±n/a nM |
|---|
| Citation |  Burgey, CS; Robinson, KA; Lyle, TA; Sanderson, PE; Lewis, SD; Lucas, BJ; Krueger, JA; Singh, R; Miller-Stein, C; White, RB; Wong, B; Lyle, EA; Williams, PD; Coburn, CA; Dorsey, BD; Barrow, JC; Stranieri, MT; Holahan, MA; Sitko, GR; Cook, JJ; McMasters, DR; McDonough, CM; Sanders, WM; Wallace, AA; Clayton, FC; Bohn, D; Leonard, YM; Detwiler, TJ; Lynch, JJ; Yan, Y; Chen, Z; Kuo, L; Gardell, SJ; Shafer, JA; Vacca, JP Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines. J Med Chem46:461-73 (2003) [PubMed] Article Burgey, CS; Robinson, KA; Lyle, TA; Sanderson, PE; Lewis, SD; Lucas, BJ; Krueger, JA; Singh, R; Miller-Stein, C; White, RB; Wong, B; Lyle, EA; Williams, PD; Coburn, CA; Dorsey, BD; Barrow, JC; Stranieri, MT; Holahan, MA; Sitko, GR; Cook, JJ; McMasters, DR; McDonough, CM; Sanders, WM; Wallace, AA; Clayton, FC; Bohn, D; Leonard, YM; Detwiler, TJ; Lynch, JJ; Yan, Y; Chen, Z; Kuo, L; Gardell, SJ; Shafer, JA; Vacca, JP Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines. J Med Chem46:461-73 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Serine protease 1 |
|---|
| Name: | Serine protease 1 |
|---|
| Synonyms: | Alpha-trypsin chain 1 | Alpha-trypsin chain 2 | Beta-trypsin | Cationic trypsinogen | PRSS1 | Serine protease 1 | TRP1 | TRY1 | TRY1_HUMAN | TRYP1 | Thrombin & trypsin | Trypsin | Trypsin I | Trypsin-1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 26557.80 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P07477 |
|---|
| Residue: | 247 |
|---|
| Sequence: | MNPLLILTFVAAALAAPFDDDDKIVGGYNCEENSVPYQVSLNSGYHFCGGSLINEQWVVS
AGHCYKSRIQVRLGEHNIEVLEGNEQFINAAKIIRHPQYDRKTLNNDIMLIKLSSRAVIN
ARVSTISLPTAPPATGTKCLISGWGNTASSGADYPDELQCLDAPVLSQAKCEASYPGKIT
SNMFCVGFLEGGKDSCQGDSGGPVVCNGQLQGVVSWGDGCAQKNKPGVYTKVYNYVKWIK
NTIAANS
|
|
|
|---|
| BDBM50123503 |
|---|
| n/a |
|---|
| Name | BDBM50123503 |
|---|
| Synonyms: | 2-[3-(2,2-Difluoro-2-pyridin-2-yl-ethylamino)-6-methyl-2-oxo-2H-pyrazin-1-yl]-N-(3-fluoro-4-methyl-pyridin-2-ylmethyl)-acetamide | CHEMBL143824 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H21F3N6O2 |
|---|
| Mol. Mass. | 446.4256 |
|---|
| SMILES | Cc1ccnc(CNC(=O)Cn2c(C)cnc(NCC(F)(F)c3ccccn3)c2=O)c1F |
|---|
| Structure | 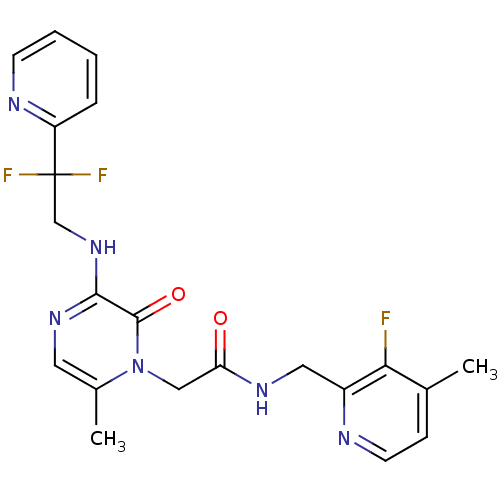
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Burgey, CS; Robinson, KA; Lyle, TA; Sanderson, PE; Lewis, SD; Lucas, BJ; Krueger, JA; Singh, R; Miller-Stein, C; White, RB; Wong, B; Lyle, EA; Williams, PD; Coburn, CA; Dorsey, BD; Barrow, JC; Stranieri, MT; Holahan, MA; Sitko, GR; Cook, JJ; McMasters, DR; McDonough, CM; Sanders, WM; Wallace, AA; Clayton, FC; Bohn, D; Leonard, YM; Detwiler, TJ; Lynch, JJ; Yan, Y; Chen, Z; Kuo, L; Gardell, SJ; Shafer, JA; Vacca, JP Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines. J Med Chem46:461-73 (2003) [PubMed] Article
Burgey, CS; Robinson, KA; Lyle, TA; Sanderson, PE; Lewis, SD; Lucas, BJ; Krueger, JA; Singh, R; Miller-Stein, C; White, RB; Wong, B; Lyle, EA; Williams, PD; Coburn, CA; Dorsey, BD; Barrow, JC; Stranieri, MT; Holahan, MA; Sitko, GR; Cook, JJ; McMasters, DR; McDonough, CM; Sanders, WM; Wallace, AA; Clayton, FC; Bohn, D; Leonard, YM; Detwiler, TJ; Lynch, JJ; Yan, Y; Chen, Z; Kuo, L; Gardell, SJ; Shafer, JA; Vacca, JP Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines. J Med Chem46:461-73 (2003) [PubMed] Article