| Reaction Details |
|---|
 | Report a problem with these data |
| Target | DNA polymerase I, thermostable |
|---|
| Ligand | BDBM50123623 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_209729 (CHEMBL816427) |
|---|
| IC50 | 5600±n/a nM |
|---|
| Citation |  Kim, MY; Duan, W; Gleason-Guzman, M; Hurley, LH Design, synthesis, and biological evaluation of a series of fluoroquinoanthroxazines with contrasting dual mechanisms of action against topoisomerase II and G-quadruplexes. J Med Chem46:571-83 (2003) [PubMed] Article Kim, MY; Duan, W; Gleason-Guzman, M; Hurley, LH Design, synthesis, and biological evaluation of a series of fluoroquinoanthroxazines with contrasting dual mechanisms of action against topoisomerase II and G-quadruplexes. J Med Chem46:571-83 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| DNA polymerase I, thermostable |
|---|
| Name: | DNA polymerase I, thermostable |
|---|
| Synonyms: | DNA polymerase I, thermostable | DPO1_THEAQ | Taq polymerase 1 | pol1 | polA |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 93906.95 |
|---|
| Organism: | Thermus aquaticus |
|---|
| Description: | ChEMBL_1289218 |
|---|
| Residue: | 832 |
|---|
| Sequence: | MRGMLPLFEPKGRVLLVDGHHLAYRTFHALKGLTTSRGEPVQAVYGFAKSLLKALKEDGD
AVIVVFDAKAPSFRHEAYGGYKAGRAPTPEDFPRQLALIKELVDLLGLARLEVPGYEADD
VLASLAKKAEKEGYEVRILTADKDLYQLLSDRIHVLHPEGYLITPAWLWEKYGLRPDQWA
DYRALTGDESDNLPGVKGIGEKTARKLLEEWGSLEALLKNLDRLKPAIREKILAHMDDLK
LSWDLAKVRTDLPLEVDFAKRREPDRERLRAFLERLEFGSLLHEFGLLESPKALEEAPWP
PPEGAFVGFVLSRKEPMWADLLALAAARGGRVHRAPEPYKALRDLKEARGLLAKDLSVLA
LREGLGLPPGDDPMLLAYLLDPSNTTPEGVARRYGGEWTEEAGERAALSERLFANLWGRL
EGEERLLWLYREVERPLSAVLAHMEATGVRLDVAYLRALSLEVAEEIARLEAEVFRLAGH
PFNLNSRDQLERVLFDELGLPAIGKTEKTGKRSTSAAVLEALREAHPIVEKILQYRELTK
LKSTYIDPLPDLIHPRTGRLHTRFNQTATATGRLSSSDPNLQNIPVRTPLGQRIRRAFIA
EEGWLLVALDYSQIELRVLAHLSGDENLIRVFQEGRDIHTETASWMFGVPREAVDPLMRR
AAKTINFGVLYGMSAHRLSQELAIPYEEAQAFIERYFQSFPKVRAWIEKTLEEGRRRGYV
ETLFGRRRYVPDLEARVKSVREAAERMAFNMPVQGTAADLMKLAMVKLFPRLEEMGARML
LQVHDELVLEAPKERAEAVARLAKEVMEGVYPLAVPLEVEVGIGEDWLSAKE
|
|
|
|---|
| BDBM50123623 |
|---|
| n/a |
|---|
| Name | BDBM50123623 |
|---|
| Synonyms: | (R)-13-(3-aminopyrrolidin-1-yl)-12-fluoro-10-oxo-10H-naphtho[1,2-c]pyrido[3,2,1-kl]phenoxazine-9-carboxylic acid | 13-[3-amino-(3R)-tetrahydro-1H-1-pyrrolyl]-12-fluoro-10-oxo-10H-naphtho[1,2-c]pyrido[3,2,1-kl]phenoxazine-9-carboxylic acid | CHEMBL346068 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H20FN3O4 |
|---|
| Mol. Mass. | 481.4745 |
|---|
| SMILES | N[C@@H]1CCN(C1)c1c(F)cc2c3c1oc1c4ccc5ccccc5c4ccc1n3cc(C(O)=O)c2=O |
|---|
| Structure | 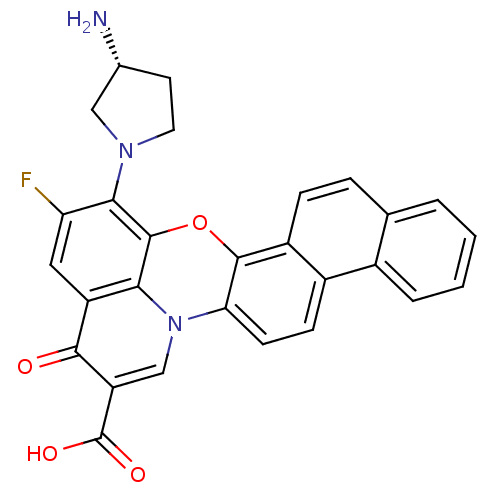
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kim, MY; Duan, W; Gleason-Guzman, M; Hurley, LH Design, synthesis, and biological evaluation of a series of fluoroquinoanthroxazines with contrasting dual mechanisms of action against topoisomerase II and G-quadruplexes. J Med Chem46:571-83 (2003) [PubMed] Article
Kim, MY; Duan, W; Gleason-Guzman, M; Hurley, LH Design, synthesis, and biological evaluation of a series of fluoroquinoanthroxazines with contrasting dual mechanisms of action against topoisomerase II and G-quadruplexes. J Med Chem46:571-83 (2003) [PubMed] Article