

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Prothrombin | ||
| Ligand | BDBM50126528 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_208514 | ||
| Ki | 0.023000±n/a nM | ||
| Citation |  Boatman, PD; Urban, J; Nguyen, M; Qabar, M; Kahn, M High-throughput synthesis and optimization of thrombin inhibitors via urazole alpha-addition and Michael addition. Bioorg Med Chem Lett13:1445-9 (2003) [PubMed] Boatman, PD; Urban, J; Nguyen, M; Qabar, M; Kahn, M High-throughput synthesis and optimization of thrombin inhibitors via urazole alpha-addition and Michael addition. Bioorg Med Chem Lett13:1445-9 (2003) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Prothrombin | |||
| Name: | Prothrombin | ||
| Synonyms: | Activation peptide fragment 1 | Activation peptide fragment 2 | Coagulation factor II | F2 | Prothrombin precursor | THRB_HUMAN | Thrombin heavy chain | Thrombin light chain | ||
| Type: | Protein | ||
| Mol. Mass.: | 70029.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P00734 | ||
| Residue: | 622 | ||
| Sequence: |
| ||
| BDBM50126528 | |||
| n/a | |||
| Name | BDBM50126528 | ||
| Synonyms: | 2-{5-[(4-Amino-cyclohexylmethyl)-carbamoyl]-1,3-dioxo-5,8-dihydro-[1,2,4]triazolo[1,2-a]pyridazin-2-yl}-3-(4-bromo-phenylsulfanyl)-propionic acid methyl ester | CHEMBL36012 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C24H30BrN5O5S | ||
| Mol. Mass. | 580.495 | ||
| SMILES | COC(=O)C(CSc1ccc(Br)cc1)n1c(=O)n2CC=CC(C(=O)NC[C@H]3CC[C@H](N)CC3)n2c1=O |wU:26.26,wD:29.30,c:20,(-5.83,6.53,;-5.06,5.2,;-3.52,5.2,;-2.75,6.53,;-2.75,3.87,;-3.52,2.54,;-5.06,2.54,;-5.83,1.21,;-5.06,-.12,;-5.83,-1.45,;-7.37,-1.45,;-8.14,-2.8,;-8.14,-.12,;-7.37,1.21,;-1.21,3.87,;-.3,5.11,;-.76,6.57,;1.17,4.64,;2.5,5.41,;3.85,4.64,;3.85,3.08,;2.5,2.31,;2.5,.77,;1.17,,;3.85,,;5.19,.77,;6.52,,;7.85,.77,;9.18,-.01,;9.18,-1.55,;10.51,-2.32,;7.84,-2.32,;6.52,-1.55,;1.17,3.08,;-.3,2.61,;-.77,1.14,)| | ||
| Structure | 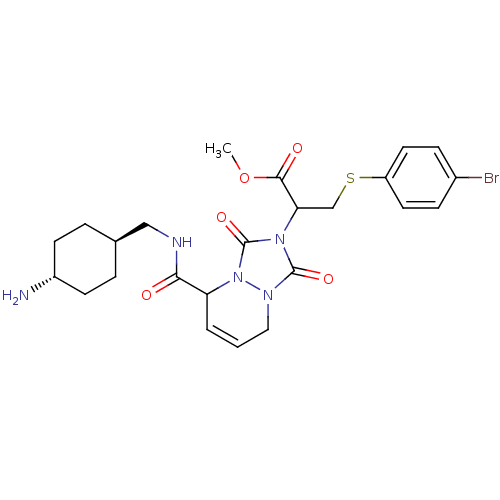
| ||