| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peptide deformylase, mitochondrial |
|---|
| Ligand | BDBM50132672 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_154318 |
|---|
| IC50 | 8390.0±n/a nM |
|---|
| Citation |  Takayama, W; Shirasaki, Y; Sakai, Y; Nakajima, E; Fujita, S; Sakamoto-Mizutani, K; Inoue, J Synthesis and PDF inhibitory activities of novel benzothiazolylidenehydroxamic acid derivatives. Bioorg Med Chem Lett13:3273-6 (2003) [PubMed] Takayama, W; Shirasaki, Y; Sakai, Y; Nakajima, E; Fujita, S; Sakamoto-Mizutani, K; Inoue, J Synthesis and PDF inhibitory activities of novel benzothiazolylidenehydroxamic acid derivatives. Bioorg Med Chem Lett13:3273-6 (2003) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peptide deformylase, mitochondrial |
|---|
| Name: | Peptide deformylase, mitochondrial |
|---|
| Synonyms: | DEFM_HUMAN | PDF | PDF1A | Peptide deformylase mitochondrial | Peptide deformylase, mitochondrial | Polypeptide deformylase |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 27023.25 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_154318 |
|---|
| Residue: | 243 |
|---|
| Sequence: | MARLWGALSLWPLWAAVPWGGAAAVGVRACSSTAAPDGVEGPALRRSYWRHLRRLVLGPP
EPPFSHVCQVGDPVLRGVAAPVERAQLGGPELQRLTQRLVQVMRRRRCVGLSAPQLGVPR
QVLALELPEALCRECPPRQRALRQMEPFPLRVFVNPSLRVLDSRLVTFPEGCESVAGFLA
CVPRFQAVQISGLDPNGEQVVWQASGWAARIIQHEMDHLQGCLFIDKMDSRTFTNVYWMK
VND
|
|
|
|---|
| BDBM50132672 |
|---|
| n/a |
|---|
| Name | BDBM50132672 |
|---|
| Synonyms: | CHEMBL419929 | N-Hydroxy-2-[3-propyl-3H-benzothiazol-(2Z)-ylidene]-acetamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C12H14N2O2S |
|---|
| Mol. Mass. | 250.317 |
|---|
| SMILES | CCCN1\C(Sc2ccccc12)=C\C(=O)NO |
|---|
| Structure | 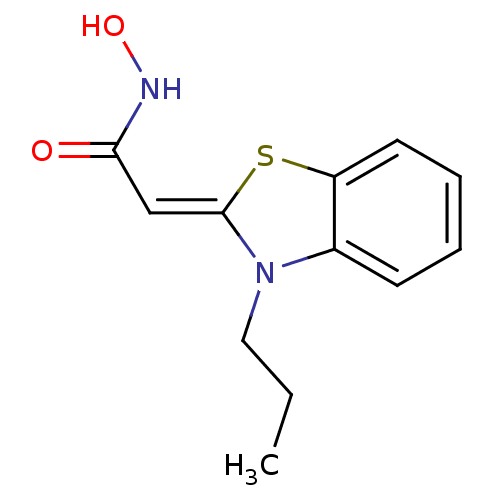
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Takayama, W; Shirasaki, Y; Sakai, Y; Nakajima, E; Fujita, S; Sakamoto-Mizutani, K; Inoue, J Synthesis and PDF inhibitory activities of novel benzothiazolylidenehydroxamic acid derivatives. Bioorg Med Chem Lett13:3273-6 (2003) [PubMed]
Takayama, W; Shirasaki, Y; Sakai, Y; Nakajima, E; Fujita, S; Sakamoto-Mizutani, K; Inoue, J Synthesis and PDF inhibitory activities of novel benzothiazolylidenehydroxamic acid derivatives. Bioorg Med Chem Lett13:3273-6 (2003) [PubMed]