| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mitogen-activated protein kinase 8 |
|---|
| Ligand | BDBM50136008 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_216826 (CHEMBL816405) |
|---|
| IC50 | 448.0±n/a nM |
|---|
| Citation |  Gingrich, DE; Reddy, DR; Iqbal, MA; Singh, J; Aimone, LD; Angeles, TS; Albom, M; Yang, S; Ator, MA; Meyer, SL; Robinson, C; Ruggeri, BA; Dionne, CA; Vaught, JL; Mallamo, JP; Hudkins, RL A new class of potent vascular endothelial growth factor receptor tyrosine kinase inhibitors: structure-activity relationships for a series of 9-alkoxymethyl-12-(3-hydroxypropyl)indeno[2,1-a]pyrrolo[3,4-c]carbazole-5-ones and the identification of CEP-5214 and its dimethylglycine ester prodrug clin J Med Chem46:5375-88 (2003) [PubMed] Article Gingrich, DE; Reddy, DR; Iqbal, MA; Singh, J; Aimone, LD; Angeles, TS; Albom, M; Yang, S; Ator, MA; Meyer, SL; Robinson, C; Ruggeri, BA; Dionne, CA; Vaught, JL; Mallamo, JP; Hudkins, RL A new class of potent vascular endothelial growth factor receptor tyrosine kinase inhibitors: structure-activity relationships for a series of 9-alkoxymethyl-12-(3-hydroxypropyl)indeno[2,1-a]pyrrolo[3,4-c]carbazole-5-ones and the identification of CEP-5214 and its dimethylglycine ester prodrug clin J Med Chem46:5375-88 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mitogen-activated protein kinase 8 |
|---|
| Name: | Mitogen-activated protein kinase 8 |
|---|
| Synonyms: | JNK-46 | JNK1 | JNK1-alpha-1 | MAPK8 | MK08_HUMAN | Mitogen-Activated Protein Kinase 8 (JNK1) | PRKM8 | SAPK1 | SAPK1C | Stress-activated protein kinase JNK1 | c-Jun N-terminal kinase 1 | c-Jun N-terminal kinase 1 (JNK1) | c-Jun N-terminal kinase 1(JNK1) | c-Jun N-terminal kinase 2 (JNK2) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48297.57 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | JNK-1 was purchased from Upstate Cell Signaling Solutions (formerly Upstate Biotechnology). |
|---|
| Residue: | 427 |
|---|
| Sequence: | MSRSKRDNNFYSVEIGDSTFTVLKRYQNLKPIGSGAQGIVCAAYDAILERNVAIKKLSRP
FQNQTHAKRAYRELVLMKCVNHKNIIGLLNVFTPQKSLEEFQDVYIVMELMDANLCQVIQ
MELDHERMSYLLYQMLCGIKHLHSAGIIHRDLKPSNIVVKSDCTLKILDFGLARTAGTSF
MMTPYVVTRYYRAPEVILGMGYKENVDLWSVGCIMGEMVCHKILFPGRDYIDQWNKVIEQ
LGTPCPEFMKKLQPTVRTYVENRPKYAGYSFEKLFPDVLFPADSEHNKLKASQARDLLSK
MLVIDASKRISVDEALQHPYINVWYDPSEAEAPPPKIPDKQLDEREHTIEEWKELIYKEV
MDLEERTKNGVIRGQPSPLGAAVINGSQHPSSSSSVNDVSSMSTDPTLASDTDSSLEAAA
GPLGCCR
|
|
|
|---|
| BDBM50136008 |
|---|
| n/a |
|---|
| Name | BDBM50136008 |
|---|
| Synonyms: | 12-(3-hydroxypropyl)-9-isopropoxymethyl-6,7,12,13-tetrahydro-5H-indeno[2,1-a]pyrrolo[4,3-c]carbazol-5-one | CHEMBL150894 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H28N2O3 |
|---|
| Mol. Mass. | 440.5335 |
|---|
| SMILES | CC(C)OCc1ccc2n(CCCO)c3c4Cc5ccccc5-c4c4C(=O)NCc4c3c2c1 |
|---|
| Structure | 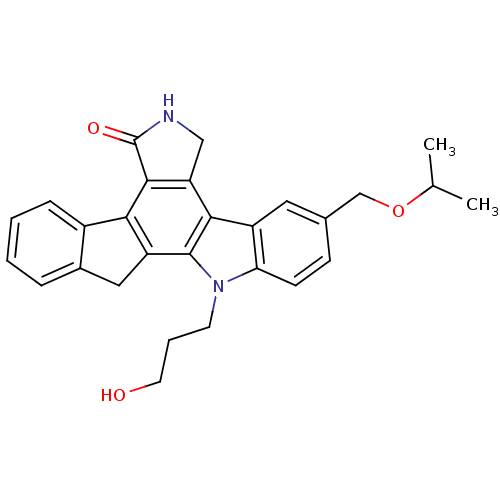
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Gingrich, DE; Reddy, DR; Iqbal, MA; Singh, J; Aimone, LD; Angeles, TS; Albom, M; Yang, S; Ator, MA; Meyer, SL; Robinson, C; Ruggeri, BA; Dionne, CA; Vaught, JL; Mallamo, JP; Hudkins, RL A new class of potent vascular endothelial growth factor receptor tyrosine kinase inhibitors: structure-activity relationships for a series of 9-alkoxymethyl-12-(3-hydroxypropyl)indeno[2,1-a]pyrrolo[3,4-c]carbazole-5-ones and the identification of CEP-5214 and its dimethylglycine ester prodrug clin J Med Chem46:5375-88 (2003) [PubMed] Article
Gingrich, DE; Reddy, DR; Iqbal, MA; Singh, J; Aimone, LD; Angeles, TS; Albom, M; Yang, S; Ator, MA; Meyer, SL; Robinson, C; Ruggeri, BA; Dionne, CA; Vaught, JL; Mallamo, JP; Hudkins, RL A new class of potent vascular endothelial growth factor receptor tyrosine kinase inhibitors: structure-activity relationships for a series of 9-alkoxymethyl-12-(3-hydroxypropyl)indeno[2,1-a]pyrrolo[3,4-c]carbazole-5-ones and the identification of CEP-5214 and its dimethylglycine ester prodrug clin J Med Chem46:5375-88 (2003) [PubMed] Article