

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Potassium voltage-gated channel subfamily H member 2 | ||
| Ligand | BDBM50137271 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_158546 | ||
| Ki | 5700±n/a nM | ||
| Citation |  Parmee, ER; He, J; Mastracchio, A; Edmondson, SD; Colwell, L; Eiermann, G; Feeney, WP; Habulihaz, B; He, H; Kilburn, R; Leiting, B; Lyons, K; Marsilio, F; Patel, RA; Petrov, A; Di Salvo, J; Wu, JK; Thornberry, NA; Weber, AE 4-Amino cyclohexylglycine analogues as potent dipeptidyl peptidase IV inhibitors. Bioorg Med Chem Lett14:43-6 (2003) [PubMed] Parmee, ER; He, J; Mastracchio, A; Edmondson, SD; Colwell, L; Eiermann, G; Feeney, WP; Habulihaz, B; He, H; Kilburn, R; Leiting, B; Lyons, K; Marsilio, F; Patel, RA; Petrov, A; Di Salvo, J; Wu, JK; Thornberry, NA; Weber, AE 4-Amino cyclohexylglycine analogues as potent dipeptidyl peptidase IV inhibitors. Bioorg Med Chem Lett14:43-6 (2003) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Potassium voltage-gated channel subfamily H member 2 | |||
| Name: | Potassium voltage-gated channel subfamily H member 2 | ||
| Synonyms: | 1,3-beta-glucan synthase component GLS2 | Cytochrome P450 3A4 | ERG | ERG1 | Eag-related protein 1 | Ether a-go-go related gene potassium channel (hERG) | Ether-a-go-go-related gene (HERG) | Ether-a-go-go-related gene potassium channel (hERG) | Ether-a-go-go-related gene potassium channel 1 | Ether-a-go-go-related gene potassium channel 1 (HERG) | Ether-a-go-go-related gene potassium channel 1 (hERG1) | Ether-a-go-go-related protein (hERG) | Ether-a-go-go-related protein 1 | Ether-a-go-go-related protein 1 (HERG) | H-ERG | HERG | KCNH2 | KCNH2_HUMAN | Potassium voltage-gated channel subfamily H member 2 (hERG) | Transcriptional regulator ERG | Voltage-gated potassium channel subunit Kv11.1 | eag homolog | hERG Potassium Channel 1 | putative potassium channel subunit | ||
| Type: | Multi-pass membrane protein | ||
| Mol. Mass.: | 126672.65 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q12809 | ||
| Residue: | 1159 | ||
| Sequence: |
| ||
| BDBM50137271 | |||
| n/a | |||
| Name | BDBM50137271 | ||
| Synonyms: | CHEMBL23979 | N-[4-((S)-1-Amino-2-oxo-2-pyrrolidin-1-yl-ethyl)-cyclohexyl]-4-trifluoromethoxy-benzenesulfonamide | ||
| Type | Small organic molecule | ||
| Emp. Form. | C19H26F3N3O4S | ||
| Mol. Mass. | 449.488 | ||
| SMILES | [H][C@@]1(CC[C@@H](CC1)NS(=O)(=O)c1ccc(OC(F)(F)F)cc1)[C@H](N)C(=O)N1CCCC1 |wU:22.24,4.7,1.0,(4.31,-7.01,;4.31,-5.47,;4.31,-3.92,;2.98,-3.15,;1.65,-3.92,;1.65,-5.47,;2.98,-6.24,;.31,-3.13,;-1.02,-3.9,;-.25,-5.25,;-2.37,-4.68,;-1.79,-2.56,;-1.02,-1.23,;-1.79,.11,;-3.34,.11,;-4.12,1.44,;-5.67,1.44,;-5.67,-.1,;-5.67,2.97,;-7.21,1.44,;-4.11,-1.23,;-3.34,-2.56,;5.64,-6.24,;5.64,-7.78,;6.97,-5.47,;6.97,-3.92,;8.3,-6.24,;8.62,-7.74,;10.15,-7.9,;10.79,-6.5,;9.64,-5.47,)| | ||
| Structure | 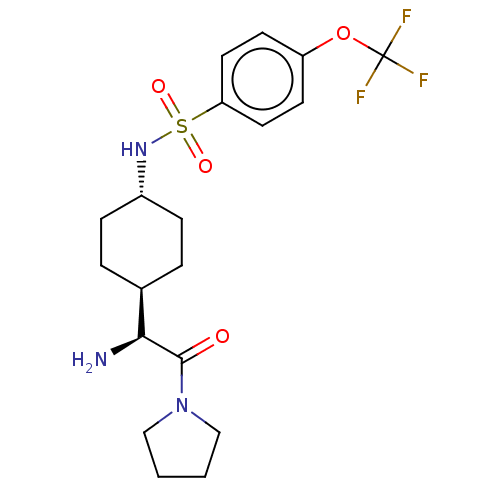
| ||