| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Plasma kallikrein |
|---|
| Ligand | BDBM50138674 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_92235 (CHEMBL703580) |
|---|
| Ki | 2720±n/a nM |
|---|
| Citation |  Wendt, MD; Rockway, TW; Geyer, A; McClellan, W; Weitzberg, M; Zhao, X; Mantei, R; Nienaber, VL; Stewart, K; Klinghofer, V; Giranda, VL Identification of novel binding interactions in the development of potent, selective 2-naphthamidine inhibitors of urokinase. Synthesis, structural analysis, and SAR of N-phenyl amide 6-substitution. J Med Chem47:303-24 (2004) [PubMed] Article Wendt, MD; Rockway, TW; Geyer, A; McClellan, W; Weitzberg, M; Zhao, X; Mantei, R; Nienaber, VL; Stewart, K; Klinghofer, V; Giranda, VL Identification of novel binding interactions in the development of potent, selective 2-naphthamidine inhibitors of urokinase. Synthesis, structural analysis, and SAR of N-phenyl amide 6-substitution. J Med Chem47:303-24 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Plasma kallikrein |
|---|
| Name: | Plasma kallikrein |
|---|
| Synonyms: | Fletcher factor | KLK3 | KLKB1 | KLKB1_HUMAN | Kallikrein | Kininogenin | Plasma kallikrein heavy chain | Plasma kallikrein light chain | Plasma prekallikrein |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 71391.16 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P03952 |
|---|
| Residue: | 638 |
|---|
| Sequence: | MILFKQATYFISLFATVSCGCLTQLYENAFFRGGDVASMYTPNAQYCQMRCTFHPRCLLF
SFLPASSINDMEKRFGCFLKDSVTGTLPKVHRTGAVSGHSLKQCGHQISACHRDIYKGVD
MRGVNFNVSKVSSVEECQKRCTSNIRCQFFSYATQTFHKAEYRNNCLLKYSPGGTPTAIK
VLSNVESGFSLKPCALSEIGCHMNIFQHLAFSDVDVARVLTPDAFVCRTICTYHPNCLFF
TFYTNVWKIESQRNVCLLKTSESGTPSSSTPQENTISGYSLLTCKRTLPEPCHSKIYPGV
DFGGEELNVTFVKGVNVCQETCTKMIRCQFFTYSLLPEDCKEEKCKCFLRLSMDGSPTRI
AYGTQGSSGYSLRLCNTGDNSVCTTKTSTRIVGGTNSSWGEWPWQVSLQVKLTAQRHLCG
GSLIGHQWVLTAAHCFDGLPLQDVWRIYSGILNLSDITKDTPFSQIKEIIIHQNYKVSEG
NHDIALIKLQAPLNYTEFQKPICLPSKGDTSTIYTNCWVTGWGFSKEKGEIQNILQKVNI
PLVTNEECQKRYQDYKITQRMVCAGYKEGGKDACKGDSGGPLVCKHNGMWRLVGITSWGE
GCARREQPGVYTKVAEYMDWILEKTQSSDGKAQMQSPA
|
|
|
|---|
| BDBM50138674 |
|---|
| n/a |
|---|
| Name | BDBM50138674 |
|---|
| Synonyms: | 6-Carbamimidoyl-naphthalene-2-carboxylic acid (3-cyclopentyloxy-phenyl)-amide | 6-[(Z)-AMINO(IMINO)METHYL]-N-[3-(CYCLOPENTYLOXY)PHENYL]-2-NAPHTHAMIDE | 6-carbamimidoyl-N-(3-(cyclopentyloxy)phenyl)-2-naphthamide | CHEMBL111575 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H23N3O2 |
|---|
| Mol. Mass. | 373.4476 |
|---|
| SMILES | NC(=N)c1ccc2cc(ccc2c1)C(=O)Nc1cccc(OC2CCCC2)c1 |
|---|
| Structure | 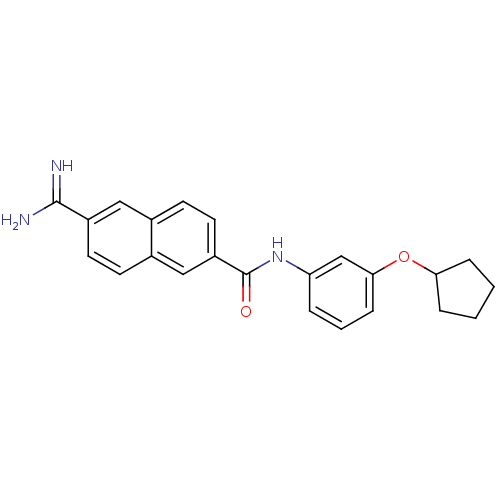
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wendt, MD; Rockway, TW; Geyer, A; McClellan, W; Weitzberg, M; Zhao, X; Mantei, R; Nienaber, VL; Stewart, K; Klinghofer, V; Giranda, VL Identification of novel binding interactions in the development of potent, selective 2-naphthamidine inhibitors of urokinase. Synthesis, structural analysis, and SAR of N-phenyl amide 6-substitution. J Med Chem47:303-24 (2004) [PubMed] Article
Wendt, MD; Rockway, TW; Geyer, A; McClellan, W; Weitzberg, M; Zhao, X; Mantei, R; Nienaber, VL; Stewart, K; Klinghofer, V; Giranda, VL Identification of novel binding interactions in the development of potent, selective 2-naphthamidine inhibitors of urokinase. Synthesis, structural analysis, and SAR of N-phenyl amide 6-substitution. J Med Chem47:303-24 (2004) [PubMed] Article