| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosine-protein kinase Fgr |
|---|
| Ligand | BDBM50151366 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_306105 (CHEMBL830879) |
|---|
| IC50 | >2.8±n/a nM |
|---|
| Citation |  Chen, P; Doweyko, AM; Norris, D; Gu, HH; Spergel, SH; Das, J; Moquin, RV; Lin, J; Wityak, J; Iwanowicz, EJ; McIntyre, KW; Shuster, DJ; Behnia, K; Chong, S; de Fex, H; Pang, S; Pitt, S; Shen, DR; Thrall, S; Stanley, P; Kocy, OR; Witmer, MR; Kanner, SB; Schieven, GL; Barrish, JC Imidazoquinoxaline Src-family kinase p56Lck inhibitors: SAR, QSAR, and the discovery of (S)-N-(2-chloro-6-methylphenyl)-2-(3-methyl-1-piperazinyl)imidazo- [1,5-a]pyrido[3,2-e]pyrazin-6-amine (BMS-279700) as a potent and orally active inhibitor with excellent in vivo antiinflammatory activity. J Med Chem47:4517-29 (2004) [PubMed] Article Chen, P; Doweyko, AM; Norris, D; Gu, HH; Spergel, SH; Das, J; Moquin, RV; Lin, J; Wityak, J; Iwanowicz, EJ; McIntyre, KW; Shuster, DJ; Behnia, K; Chong, S; de Fex, H; Pang, S; Pitt, S; Shen, DR; Thrall, S; Stanley, P; Kocy, OR; Witmer, MR; Kanner, SB; Schieven, GL; Barrish, JC Imidazoquinoxaline Src-family kinase p56Lck inhibitors: SAR, QSAR, and the discovery of (S)-N-(2-chloro-6-methylphenyl)-2-(3-methyl-1-piperazinyl)imidazo- [1,5-a]pyrido[3,2-e]pyrazin-6-amine (BMS-279700) as a potent and orally active inhibitor with excellent in vivo antiinflammatory activity. J Med Chem47:4517-29 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Tyrosine-protein kinase Fgr |
|---|
| Name: | Tyrosine-protein kinase Fgr |
|---|
| Synonyms: | FGR | FGR_HUMAN | Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog | Proto-oncogene c-Fgr | SRC | SRC2 | p55-Fgr | p58-Fgr | p58c-Fgr |
|---|
| Type: | protein |
|---|
| Mol. Mass.: | 59469.88 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P09769 |
|---|
| Residue: | 529 |
|---|
| Sequence: | MGCVFCKKLEPVATAKEDAGLEGDFRSYGAADHYGPDPTKARPASSFAHIPNYSNFSSQA
INPGFLDSGTIRGVSGIGVTLFIALYDYEARTEDDLTFTKGEKFHILNNTEGDWWEARSL
SSGKTGCIPSNYVAPVDSIQAEEWYFGKIGRKDAERQLLSPGNPQGAFLIRESETTKGAY
SLSIRDWDQTRGDHVKHYKIRKLDMGGYYITTRVQFNSVQELVQHYMEVNDGLCNLLIAP
CTIMKPQTLGLAKDAWEISRSSITLERRLGTGCFGDVWLGTWNGSTKVAVKTLKPGTMSP
KAFLEEAQVMKLLRHDKLVQLYAVVSEEPIYIVTEFMCHGSLLDFLKNPEGQDLRLPQLV
DMAAQVAEGMAYMERMNYIHRDLRAANILVGERLACKIADFGLARLIKDDEYNPCQGSKF
PIKWTAPEAALFGRFTIKSDVWSFGILLTELITKGRIPYPGMNKREVLEQVEQGYHMPCP
PGCPASLYEAMEQTWRLDPEERPTFEYLQSFLEDYFTSAEPQYQPGDQT
|
|
|
|---|
| BDBM50151366 |
|---|
| n/a |
|---|
| Name | BDBM50151366 |
|---|
| Synonyms: | (2-Chloro-6-methyl-phenyl)-[8-((S)-3-methyl-piperazin-1-yl)-2,5,9,9b-tetraaza-cyclopenta[a]naphthalen-4-yl]-amine | BMS-279700 | CHEMBL189338 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H22ClN7 |
|---|
| Mol. Mass. | 407.899 |
|---|
| SMILES | C[C@H]1CN(CCN1)c1ccc2nc(Nc3c(C)cccc3Cl)c3cncn3c2n1 |
|---|
| Structure | 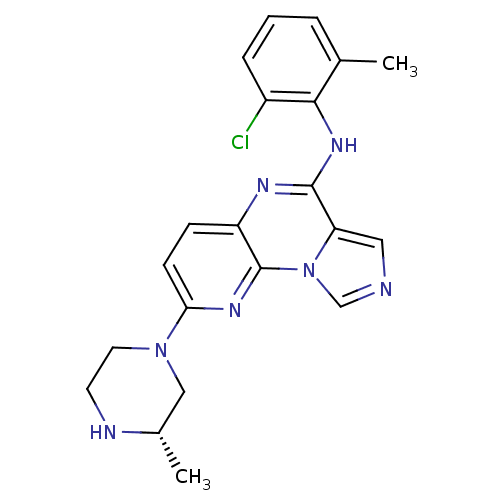
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Chen, P; Doweyko, AM; Norris, D; Gu, HH; Spergel, SH; Das, J; Moquin, RV; Lin, J; Wityak, J; Iwanowicz, EJ; McIntyre, KW; Shuster, DJ; Behnia, K; Chong, S; de Fex, H; Pang, S; Pitt, S; Shen, DR; Thrall, S; Stanley, P; Kocy, OR; Witmer, MR; Kanner, SB; Schieven, GL; Barrish, JC Imidazoquinoxaline Src-family kinase p56Lck inhibitors: SAR, QSAR, and the discovery of (S)-N-(2-chloro-6-methylphenyl)-2-(3-methyl-1-piperazinyl)imidazo- [1,5-a]pyrido[3,2-e]pyrazin-6-amine (BMS-279700) as a potent and orally active inhibitor with excellent in vivo antiinflammatory activity. J Med Chem47:4517-29 (2004) [PubMed] Article
Chen, P; Doweyko, AM; Norris, D; Gu, HH; Spergel, SH; Das, J; Moquin, RV; Lin, J; Wityak, J; Iwanowicz, EJ; McIntyre, KW; Shuster, DJ; Behnia, K; Chong, S; de Fex, H; Pang, S; Pitt, S; Shen, DR; Thrall, S; Stanley, P; Kocy, OR; Witmer, MR; Kanner, SB; Schieven, GL; Barrish, JC Imidazoquinoxaline Src-family kinase p56Lck inhibitors: SAR, QSAR, and the discovery of (S)-N-(2-chloro-6-methylphenyl)-2-(3-methyl-1-piperazinyl)imidazo- [1,5-a]pyrido[3,2-e]pyrazin-6-amine (BMS-279700) as a potent and orally active inhibitor with excellent in vivo antiinflammatory activity. J Med Chem47:4517-29 (2004) [PubMed] Article