

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Epidermal growth factor receptor | ||
| Ligand | BDBM3096 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_306181 (CHEMBL830975) | ||
| IC50 | 44000±n/a nM | ||
| Citation |  Palmer, BD; Smaill, JB; Rewcastle, GW; Dobrusin, EM; Kraker, A; Moore, CW; Steinkampf, RW; Denny, WA Structure-activity relationships for 2-anilino-6-phenylpyrido[2,3-d]pyrimidin-7(8H)-ones as inhibitors of the cellular checkpoint kinase Wee1. Bioorg Med Chem Lett15:1931-5 (2005) [PubMed] Article Palmer, BD; Smaill, JB; Rewcastle, GW; Dobrusin, EM; Kraker, A; Moore, CW; Steinkampf, RW; Denny, WA Structure-activity relationships for 2-anilino-6-phenylpyrido[2,3-d]pyrimidin-7(8H)-ones as inhibitors of the cellular checkpoint kinase Wee1. Bioorg Med Chem Lett15:1931-5 (2005) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Epidermal growth factor receptor | |||
| Name: | Epidermal growth factor receptor | ||
| Synonyms: | EGF Receptor Substrate | EGFR | EGFR_HUMAN | ERBB | ERBB1 | Epidermal Growth Factor Receptor Tyrosine Kinase | Epidermal Growth Factor Receptor erbB-1 | Epidermal Growth Factor Receptor, ErbB-1 | Epidermal growth factor receptor (EGFR) | Epidermal growth factor receptor (HER1) | HER1 | Protein-Tyrosine Kinase Erbb-1 | Proto-oncogene c-ErbB-1 | Receptor tyrosine-protein kinase ErbB-1 | Tyrosine-Protein Kinase ErbB-1 | ||
| Type: | Receptor Kinase Domain | ||
| Mol. Mass.: | 134279.59 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P00533 | ||
| Residue: | 1210 | ||
| Sequence: |
| ||
| BDBM3096 | |||
| n/a | |||
| Name | BDBM3096 | ||
| Synonyms: | 2-aminopyrido[2,3-d]pyrimidin-7(8H)-one deriv. | 6-(2,6-Dichlorophenyl)-2-[4-(2-diethylaminoethoxy)phenylamino]-8-methyl-8H-pyrido[2,3-d]pyrimidin-7-one, Dihydrochloride | 6-(2,6-dichlorophenyl)-2-({4-[2-(diethylamino)ethoxy]phenyl}amino)-8-methyl-7H,8H-pyrido[2,3-d]pyrimidin-7-one | CHEMBL49120 | D3RKN_84 | PD166285 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H27Cl2N5O2 | ||
| Mol. Mass. | 512.431 | ||
| SMILES | CCN(CC)CCOc1ccc(Nc2ncc3cc(-c4c(Cl)cccc4Cl)c(=O)n(C)c3n2)cc1 |(-8.3,-10.68,;-6.82,-11.08,;-5.73,-9.99,;-4.24,-10.39,;-3.15,-9.3,;-6.82,-8.9,;-6.82,-7.36,;-8.15,-6.59,;-8.15,-5.05,;-6.82,-4.28,;-6.82,-2.74,;-8.15,-1.97,;-8.15,-.43,;-6.82,.34,;-6.82,1.88,;-5.48,2.65,;-4.15,1.88,;-2.82,2.65,;-1.48,1.88,;-.15,2.65,;-.15,4.19,;-1.48,4.96,;1.18,4.96,;2.52,4.19,;2.52,2.65,;1.18,1.88,;1.18,.34,;-1.48,.34,;-.15,-.43,;-2.82,-.43,;-2.82,-1.97,;-4.15,.34,;-5.48,-.43,;-9.48,-2.74,;-9.48,-4.28,)| | ||
| Structure | 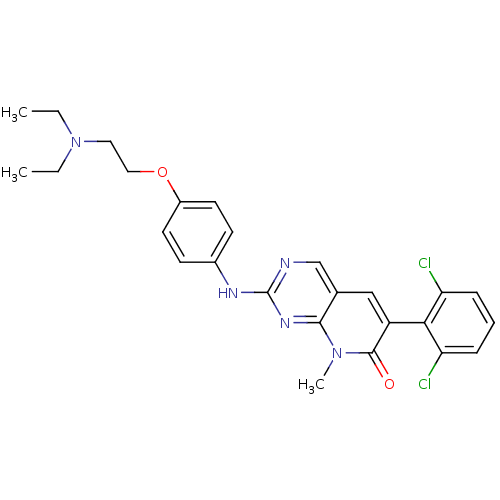
| ||