

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Wee1-like protein kinase | ||
| Ligand | BDBM50164158 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_304894 (CHEMBL829373) | ||
| IC50 | 124±n/a nM | ||
| Citation |  Palmer, BD; Smaill, JB; Rewcastle, GW; Dobrusin, EM; Kraker, A; Moore, CW; Steinkampf, RW; Denny, WA Structure-activity relationships for 2-anilino-6-phenylpyrido[2,3-d]pyrimidin-7(8H)-ones as inhibitors of the cellular checkpoint kinase Wee1. Bioorg Med Chem Lett15:1931-5 (2005) [PubMed] Article Palmer, BD; Smaill, JB; Rewcastle, GW; Dobrusin, EM; Kraker, A; Moore, CW; Steinkampf, RW; Denny, WA Structure-activity relationships for 2-anilino-6-phenylpyrido[2,3-d]pyrimidin-7(8H)-ones as inhibitors of the cellular checkpoint kinase Wee1. Bioorg Med Chem Lett15:1931-5 (2005) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Wee1-like protein kinase | |||
| Name: | Wee1-like protein kinase | ||
| Synonyms: | Serine/threonine-protein kinase WEE1 | WEE1 | WEE1 homolog (S. pombe) | WEE1_HUMAN | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 71599.12 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | ChEMBL_1445054 | ||
| Residue: | 646 | ||
| Sequence: |
| ||
| BDBM50164158 | |||
| n/a | |||
| Name | BDBM50164158 | ||
| Synonyms: | 4-{4-[6-(2,6-Dichloro-phenyl)-8-methyl-7-oxo-7,8-dihydro-pyrido[2,3-d]pyrimidin-2-ylamino]-phenyl}-butyric acid 2-dimethylamino-ethyl ester | CHEMBL179065 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H29Cl2N5O3 | ||
| Mol. Mass. | 554.468 | ||
| SMILES | CN(C)CCOC(=O)CCCc1ccc(Nc2ncc3cc(-c4c(Cl)cccc4Cl)c(=O)n(C)c3n2)cc1 |(-11.37,8.52,;-10.07,7.73,;-8.72,8.5,;-10.05,6.22,;-8.72,5.45,;-8.72,3.91,;-7.39,3.14,;-6.04,3.92,;-7.37,1.6,;-6.04,.84,;-6.04,-.7,;-4.69,-1.45,;-4.68,-3.02,;-3.35,-3.76,;-2.03,-3.01,;-.69,-3.76,;.64,-2.99,;.64,-1.45,;1.97,-.66,;3.3,-1.45,;4.63,-.66,;5.96,-1.43,;7.29,-.65,;8.62,-1.41,;8.63,-2.95,;9.95,-.65,;9.95,.9,;8.59,1.66,;7.28,.88,;5.93,1.65,;5.97,-2.99,;7.31,-3.74,;4.64,-3.76,;4.64,-5.3,;3.3,-2.99,;1.97,-3.76,;-2.02,-1.45,;-3.36,-.7,)| | ||
| Structure | 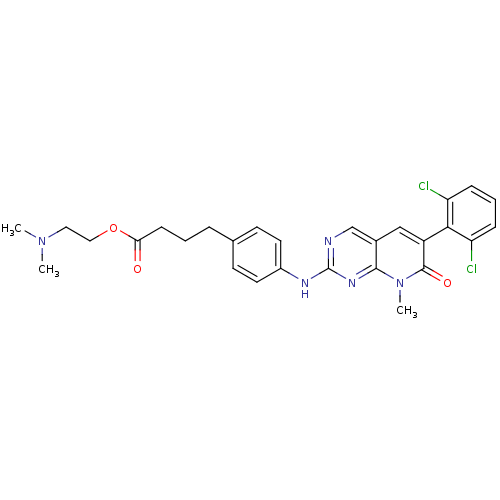
| ||