| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Ribosomal protein S6 kinase beta-1 |
|---|
| Ligand | BDBM14032 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_304643 (CHEMBL828526) |
|---|
| IC50 | 70±n/a nM |
|---|
| Citation |  Bamford, MJ; Bailey, N; Davies, S; Dean, DK; Francis, L; Panchal, TA; Parr, CA; Sehmi, S; Steadman, JG; Takle, AK; Townsend, JT; Wilson, DM (1H-imidazo[4,5-c]pyridin-2-yl)-1,2,5-oxadiazol-3-ylamine derivatives: further optimisation as highly potent and selective MSK-1-inhibitors. Bioorg Med Chem Lett15:3407-11 (2005) [PubMed] Article Bamford, MJ; Bailey, N; Davies, S; Dean, DK; Francis, L; Panchal, TA; Parr, CA; Sehmi, S; Steadman, JG; Takle, AK; Townsend, JT; Wilson, DM (1H-imidazo[4,5-c]pyridin-2-yl)-1,2,5-oxadiazol-3-ylamine derivatives: further optimisation as highly potent and selective MSK-1-inhibitors. Bioorg Med Chem Lett15:3407-11 (2005) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Ribosomal protein S6 kinase beta-1 |
|---|
| Name: | Ribosomal protein S6 kinase beta-1 |
|---|
| Synonyms: | 70 kDa ribosomal protein S6 kinase 1 | 70 kDa ribosomal protein S6 kinase 1 (P70s6k) | 70 kDa ribosomal protein kinase (p70S6K) | KS6B1_HUMAN | Protein kinase 70S6K (T412E) | RAC-alpha serine/threonine-protein kinase/Ribosomal protein S6 kinase beta-1 | RPS6KB1 | Ribosomal protein S6 kinase (P70S6K) | Ribosomal protein S6 kinase I | S6K1 | STK14A | p70 S6 ribosomal protein kinase 1 | p70S6K1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 59139.89 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | His-tagged S6K1. |
|---|
| Residue: | 525 |
|---|
| Sequence: | MRRRRRRDGFYPAPDFRDREAEDMAGVFDIDLDQPEDAGSEDELEEGGQLNESMDHGGVG
PYELGMEHCEKFEISETSVNRGPEKIRPECFELLRVLGKGGYGKVFQVRKVTGANTGKIF
AMKVLKKAMIVRNAKDTAHTKAERNILEEVKHPFIVDLIYAFQTGGKLYLILEYLSGGEL
FMQLEREGIFMEDTACFYLAEISMALGHLHQKGIIYRDLKPENIMLNHQGHVKLTDFGLC
KESIHDGTVTHTFCGTIEYMAPEILMRSGHNRAVDWWSLGALMYDMLTGAPPFTGENRKK
TIDKILKCKLNLPPYLTQEARDLLKKLLKRNAASRLGAGPGDAGEVQAHPFFRHINWEEL
LARKVEPPFKPLLQSEEDVSQFDSKFTRQTPVDSPDDSTLSESANQVFLGFTYVAPSVLE
SVKEKFSFEPKIRSPRRFIGSPRTPVSPVKFSPGDFWGRGASASTANPQTPVEYPMETSG
IEQMDVTMSGEASAPLPIRQPNSGPYKKQAFPMISKRPEHLRMNL
|
|
|
|---|
| BDBM14032 |
|---|
| n/a |
|---|
| Name | BDBM14032 |
|---|
| Synonyms: | 4-(1-ethyl-1H-imidazo[4,5-c]pyridin-2-yl)-1,2,5-oxadiazol-3-amine | 4-{1-ethyl-1H-imidazo[4,5-c]pyridin-2-yl}-1,2,5-oxadiazol-3-amine | Aminofurazanyl-azabenzimidazole 1 | CHEMBL189657 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H10N6O |
|---|
| Mol. Mass. | 230.226 |
|---|
| SMILES | CCn1c(nc2cnccc12)-c1nonc1N |
|---|
| Structure | 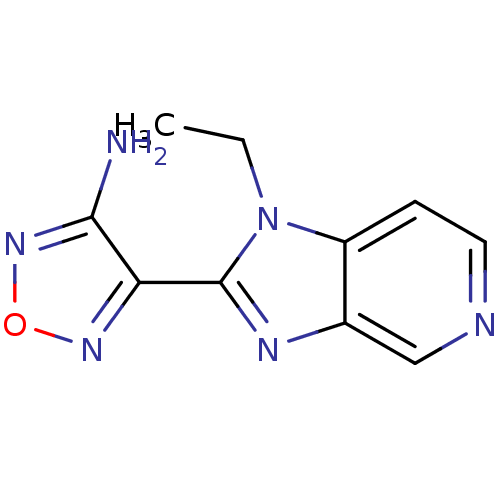
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Bamford, MJ; Bailey, N; Davies, S; Dean, DK; Francis, L; Panchal, TA; Parr, CA; Sehmi, S; Steadman, JG; Takle, AK; Townsend, JT; Wilson, DM (1H-imidazo[4,5-c]pyridin-2-yl)-1,2,5-oxadiazol-3-ylamine derivatives: further optimisation as highly potent and selective MSK-1-inhibitors. Bioorg Med Chem Lett15:3407-11 (2005) [PubMed] Article
Bamford, MJ; Bailey, N; Davies, S; Dean, DK; Francis, L; Panchal, TA; Parr, CA; Sehmi, S; Steadman, JG; Takle, AK; Townsend, JT; Wilson, DM (1H-imidazo[4,5-c]pyridin-2-yl)-1,2,5-oxadiazol-3-ylamine derivatives: further optimisation as highly potent and selective MSK-1-inhibitors. Bioorg Med Chem Lett15:3407-11 (2005) [PubMed] Article