| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Gamma-aminobutyric acid receptor subunit alpha-3/beta-3/gamma-2 |
|---|
| Ligand | BDBM50185519 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_373310 (CHEMBL853517) |
|---|
| Ki | 1.1±n/a nM |
|---|
| Citation |  Carling, RW; Russell, MG; Moore, KW; Mitchinson, A; Guiblin, A; Smith, A; Wafford, KA; Marshall, G; Atack, JR; Street, LJ 2,3,7-Trisubstituted pyrazolo[1,5-d][1,2,4]triazines: functionally selective GABAA alpha3-subtype agonists. Bioorg Med Chem Lett16:3550-4 (2006) [PubMed] Article Carling, RW; Russell, MG; Moore, KW; Mitchinson, A; Guiblin, A; Smith, A; Wafford, KA; Marshall, G; Atack, JR; Street, LJ 2,3,7-Trisubstituted pyrazolo[1,5-d][1,2,4]triazines: functionally selective GABAA alpha3-subtype agonists. Bioorg Med Chem Lett16:3550-4 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Gamma-aminobutyric acid receptor subunit alpha-3/beta-3/gamma-2 |
|---|
| Name: | Gamma-aminobutyric acid receptor subunit alpha-3/beta-3/gamma-2 |
|---|
| Synonyms: | GABA receptor alpha-3/beta-3/gamma-2 subunit | GABA-A receptor; alpha-3/beta-3/gamma-2 |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of EBI is 68410 |
|---|
| Components: | This complex has 3 components. |
|---|
| Component 1 |
| Name: | Gamma-aminobutyric acid receptor subunit gamma-2 |
|---|
| Synonyms: | GABA(A) receptor subunit gamma-2 | GABRG2 | GBRG2_HUMAN |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 54172.74 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | EBI_217 |
|---|
| Residue: | 467 |
|---|
| Sequence: | MSSPNIWSTGSSVYSTPVFSQKMTVWILLLLSLYPGFTSQKSDDDYEDYASNKTWVLTPK
VPEGDVTVILNNLLEGYDNKLRPDIGVKPTLIHTDMYVNSIGPVNAINMEYTIDIFFAQT
WYDRRLKFNSTIKVLRLNSNMVGKIWIPDTFFRNSKKADAHWITTPNRMLRIWNDGRVLY
TLRLTIDAECQLQLHNFPMDEHSCPLEFSSYGYPREEIVYQWKRSSVEVGDTRSWRLYQF
SFVGLRNTTEVVKTTSGDYVVMSVYFDLSRRMGYFTIQTYIPCTLIVVLSWVSFWINKDA
VPARTSLGITTVLTMTTLSTIARKSLPKVSYVTAMDLFVSVCFIFVFSALVEYGTLHYFV
SNRKPSKDKDKKKKNPAPTIDIRPRSATIQMNNATHLQERDEEYGYECLDGKDCASFFCC
FEDCRTGAWRHGRIHIRIAKMDSYARIFFPTAFCLFNLVYWVSYLYL
|
|
|
|---|
| Component 2 |
| Name: | Gamma-aminobutyric acid receptor subunit beta-3 |
|---|
| Synonyms: | GABA A receptor alpha-4/beta-3/gamma-2 | GABA receptor beta-3 subunit | GABA-A receptor | GABRB3 | GBRB3_HUMAN | agonist GABA site |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 54130.51 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_448071 |
|---|
| Residue: | 473 |
|---|
| Sequence: | MWGLAGGRLFGIFSAPVLVAVVCCAQSVNDPGNMSFVKETVDKLLKGYDIRLRPDFGGPP
VCVGMNIDIASIDMVSEVNMDYTLTMYFQQYWRDKRLAYSGIPLNLTLDNRVADQLWVPD
TYFLNDKKSFVHGVTVKNRMIRLHPDGTVLYGLRITTTAACMMDLRRYPLDEQNCTLEIE
SYGYTTDDIEFYWRGGDKAVTGVERIELPQFSIVEHRLVSRNVVFATGAYPRLSLSFRLK
RNIGYFILQTYMPSILITILSWVSFWINYDASAARVALGITTVLTMTTINTHLRETLPKI
PYVKAIDMYLMGCFVFVFLALLEYAFVNYIFFGRGPQRQKKLAEKTAKAKNDRSKSESNR
VDAHGNILLTSLEVHNEMNEVSGGIGDTRNSAISFDNSGIQYRKQSMPREGHGRFLGDRS
LPHKKTHLRRRSSQLKIKIPDLTDVNAIDRWSRIVFPFTFSLFNLVYWLYYVN
|
|
|
|---|
| Component 3 |
| Name: | Gamma-aminobutyric acid receptor subunit alpha-3 |
|---|
| Synonyms: | GABA A Alpha3Beta3Gamma2 | GABA A receptor alpha-3/beta-2/gamma-2 | GABA C | GABA receptor alpha-3 subunit | GABA-A receptor | GABRA3 | GBRA3_HUMAN | Gamma-aminobutyric acid receptor subunit alpha-3 | agonist GABA site |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 55175.47 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | GABA A Alpha3Beta1Gamma2 0 HUMAN::P34903 |
|---|
| Residue: | 492 |
|---|
| Sequence: | MIITQTSHCYMTSLGILFLINILPGTTGQGESRRQEPGDFVKQDIGGLSPKHAPDIPDDS
TDNITIFTRILDRLLDGYDNRLRPGLGDAVTEVKTDIYVTSFGPVSDTDMEYTIDVFFRQ
TWHDERLKFDGPMKILPLNNLLASKIWTPDTFFHNGKKSVAHNMTTPNKLLRLVDNGTLL
YTMRLTIHAECPMHLEDFPMDVHACPLKFGSYAYTTAEVVYSWTLGKNKSVEVAQDGSRL
NQYDLLGHVVGTEIIRSSTGEYVVMTTHFHLKRKIGYFVIQTYLPCIMTVILSQVSFWLN
RESVPARTVFGVTTVLTMTTLSISARNSLPKVAYATAMDWFIAVCYAFVFSALIEFATVN
YFTKRSWAWEGKKVPEALEMKKKTPAAPAKKTSTTFNIVGTTYPINLAKDTEFSTISKGA
APSASSTPTIIASPKATYVQDSPTETKTYNSVSKVDKISRIIFPVLFAIFNLVYWATYVN
RESAIKGMIRKQ
|
|
|
|---|
| BDBM50185519 |
|---|
| n/a |
|---|
| Name | BDBM50185519 |
|---|
| Synonyms: | 3-(2-fluorophenyl)-2-((2-methyl-2H-1,2,4-triazol-3-yl)methoxy)-7-(pyridin-3-yl)pyrazolo[1,5-d][1,2,4]triazine | CHEMBL210400 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H15FN8O |
|---|
| Mol. Mass. | 402.3845 |
|---|
| SMILES | Cn1ncnc1COc1nn2c(nncc2c1-c1ccccc1F)-c1cccnc1 |(6.61,-18.95,;5.09,-18.73,;4.02,-19.84,;2.63,-19.16,;2.85,-17.63,;4.38,-17.36,;5.15,-16.03,;4.39,-14.7,;5.16,-13.37,;6.7,-13.21,;7.02,-11.7,;8.37,-10.93,;8.36,-9.38,;7.02,-8.62,;5.7,-9.39,;5.69,-10.92,;4.54,-11.95,;3.2,-11.19,;3.21,-9.65,;1.87,-8.89,;.54,-9.66,;.55,-11.21,;1.89,-11.97,;1.9,-13.51,;9.7,-11.7,;9.7,-13.24,;11.03,-14.01,;12.37,-13.24,;12.36,-11.69,;11.03,-10.93,)| |
|---|
| Structure | 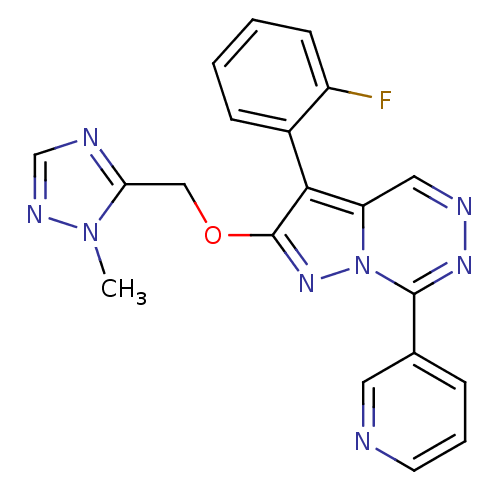
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Carling, RW; Russell, MG; Moore, KW; Mitchinson, A; Guiblin, A; Smith, A; Wafford, KA; Marshall, G; Atack, JR; Street, LJ 2,3,7-Trisubstituted pyrazolo[1,5-d][1,2,4]triazines: functionally selective GABAA alpha3-subtype agonists. Bioorg Med Chem Lett16:3550-4 (2006) [PubMed] Article
Carling, RW; Russell, MG; Moore, KW; Mitchinson, A; Guiblin, A; Smith, A; Wafford, KA; Marshall, G; Atack, JR; Street, LJ 2,3,7-Trisubstituted pyrazolo[1,5-d][1,2,4]triazines: functionally selective GABAA alpha3-subtype agonists. Bioorg Med Chem Lett16:3550-4 (2006) [PubMed] Article