

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Metabotropic glutamate receptor 1 | ||
| Ligand | BDBM50197254 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_453190 (CHEMBL902340) | ||
| Ki | 3±n/a nM | ||
| Citation |  Owen, DR; Dodd, PG; Gayton, S; Greener, BS; Harbottle, GW; Mantell, SJ; Maw, GN; Osborne, SA; Rees, H; Ringer, TJ; Rodriguez-Lens, M; Smith, GF Structure-activity relationships of novel non-competitive mGluR1 antagonists: a potential treatment for chronic pain. Bioorg Med Chem Lett17:486-90 (2007) [PubMed] Article Owen, DR; Dodd, PG; Gayton, S; Greener, BS; Harbottle, GW; Mantell, SJ; Maw, GN; Osborne, SA; Rees, H; Ringer, TJ; Rodriguez-Lens, M; Smith, GF Structure-activity relationships of novel non-competitive mGluR1 antagonists: a potential treatment for chronic pain. Bioorg Med Chem Lett17:486-90 (2007) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Metabotropic glutamate receptor 1 | |||
| Name: | Metabotropic glutamate receptor 1 | ||
| Synonyms: | GRM1_RAT | Gprc1a | Grm1 | Metabotropic Glutamate 1a | Metabotropic glutamate receptor | Metabotropic glutamate receptor 1 | Mglur1 | metabotropic glutamate 1 | metabotropic glutamate 1/5-D | metabotropic glutamate 1/DA | metabotropic glutamate receptor 1 isoform alpha precursor | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 133240.47 | ||
| Organism: | RAT | ||
| Description: | metabotropic glutamate 1/2 0 RAT::P23385 | ||
| Residue: | 1199 | ||
| Sequence: |
| ||
| BDBM50197254 | |||
| n/a | |||
| Name | BDBM50197254 | ||
| Synonyms: | CHEMBL246250 | N-((1r,4r)-4-methylcyclohexyl)-5-(piperidin-1-yl)pyrazine-2-carboxamide | ||
| Type | Small organic molecule | ||
| Emp. Form. | C17H26N4O | ||
| Mol. Mass. | 302.4145 | ||
| SMILES | C[C@H]1CC[C@@H](CC1)NC(=O)c1cnc(cn1)N1CCCCC1 |wU:4.7,wD:1.0,(7.01,-30.31,;5.68,-31.07,;4.34,-30.31,;3.01,-31.08,;3.02,-32.61,;4.35,-33.38,;5.68,-32.61,;1.69,-33.39,;.35,-32.62,;.35,-31.08,;-.98,-33.4,;-.97,-34.95,;-2.31,-35.72,;-3.64,-34.95,;-3.64,-33.4,;-2.31,-32.63,;-4.97,-35.72,;-6.31,-34.95,;-7.64,-35.71,;-7.65,-37.25,;-6.31,-38.02,;-4.97,-37.26,)| | ||
| Structure | 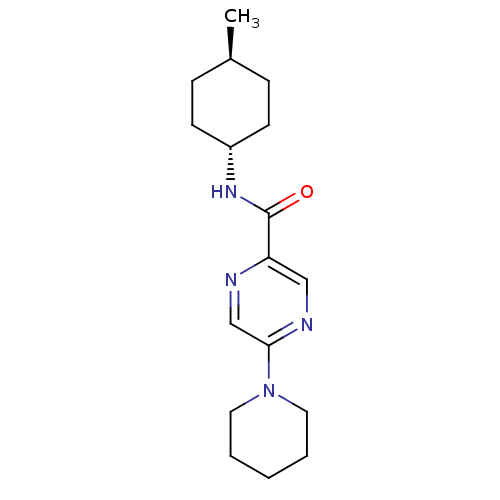
| ||