| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Integrin alpha-V |
|---|
| Ligand | BDBM50260254 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_504851 (CHEMBL983998) |
|---|
| IC50 | 1540±n/a nM |
|---|
| Citation |  DiCara, D; Rapisarda, C; Sutcliffe, JL; Violette, SM; Weinreb, PH; Hart, IR; Howard, MJ; Marshall, JF Structure-function analysis of Arg-Gly-Asp helix motifs in alpha v beta 6 integrin ligands. J Biol Chem282:9657-65 (2007) [PubMed] Article DiCara, D; Rapisarda, C; Sutcliffe, JL; Violette, SM; Weinreb, PH; Hart, IR; Howard, MJ; Marshall, JF Structure-function analysis of Arg-Gly-Asp helix motifs in alpha v beta 6 integrin ligands. J Biol Chem282:9657-65 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Integrin alpha-V |
|---|
| Name: | Integrin alpha-V |
|---|
| Synonyms: | ITAV_HUMAN | ITGAV | Integrin alpha-V/alpha-5 | MSK8 | VNRA | VTNR | Vitronectin receptor | Vitronectin receptor subunit alpha |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 116024.92 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_582336 |
|---|
| Residue: | 1048 |
|---|
| Sequence: | MAFPPRRRLRLGPRGLPLLLSGLLLPLCRAFNLDVDSPAEYSGPEGSYFGFAVDFFVPSA
SSRMFLLVGAPKANTTQPGIVEGGQVLKCDWSSTRRCQPIEFDATGNRDYAKDDPLEFKS
HQWFGASVRSKQDKILACAPLYHWRTEMKQEREPVGTCFLQDGTKTVEYAPCRSQDIDAD
GQGFCQGGFSIDFTKADRVLLGGPGSFYWQGQLISDQVAEIVSKYDPNVYSIKYNNQLAT
RTAQAIFDDSYLGYSVAVGDFNGDGIDDFVSGVPRAARTLGMVYIYDGKNMSSLYNFTGE
QMAAYFGFSVAATDINGDDYADVFIGAPLFMDRGSDGKLQEVGQVSVSLQRASGDFQTTK
LNGFEVFARFGSAIAPLGDLDQDGFNDIAIAAPYGGEDKKGIVYIFNGRSTGLNAVPSQI
LEGQWAARSMPPSFGYSMKGATDIDKNGYPDLIVGAFGVDRAILYRARPVITVNAGLEVY
PSILNQDNKTCSLPGTALKVSCFNVRFCLKADGKGVLPRKLNFQVELLLDKLKQKGAIRR
ALFLYSRSPSHSKNMTISRGGLMQCEELIAYLRDESEFRDKLTPITIFMEYRLDYRTAAD
TTGLQPILNQFTPANISRQAHILLDCGEDNVCKPKLEVSVDSDQKKIYIGDDNPLTLIVK
AQNQGEGAYEAELIVSIPLQADFIGVVRNNEALARLSCAFKTENQTRQVVCDLGNPMKAG
TQLLAGLRFSVHQQSEMDTSVKFDLQIQSSNLFDKVSPVVSHKVDLAVLAAVEIRGVSSP
DHVFLPIPNWEHKENPETEEDVGPVVQHIYELRNNGPSSFSKAMLHLQWPYKYNNNTLLY
ILHYDIDGPMNCTSDMEINPLRIKISSLQTTEKNDTVAGQGERDHLITKRDLALSEGDIH
TLGCGVAQCLKIVCQVGRLDRGKSAILYVKSLLWTETFMNKENQNHSYSLKSSASFNVIE
FPYKNLPIEDITNSTLVTTNVTWGIQPAPMPVPVWVIILAVLAGLLLLAVLVFVMYRMGF
FKRVRPPQEEQEREQLQPHENGEGNSET
|
|
|
|---|
| BDBM50260254 |
|---|
| n/a |
|---|
| Name | BDBM50260254 |
|---|
| Synonyms: | CHEMBL446897 | NAVPNLRGDLQVLAQKVART |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C93H163N31O28 |
|---|
| Mol. Mass. | 2163.4802 |
|---|
| SMILES | [#6]-[#6](-[#6])-[#6]-[#6@H](-[#7]-[#6](=O)-[#6@@H](-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#6])-[#6])-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#8])=O)-[#7]-[#6](=O)-[#6]-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#6])-[#6])-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@@H]-1-[#6]-[#6]-[#6]-[#7]-1-[#6](=O)-[#6@@H](-[#7]-[#6](=O)-[#6@H](-[#6])-[#7]-[#6](=O)-[#6@@H](-[#7])-[#6]-[#6](-[#7])=O)-[#6](-[#6])-[#6])-[#6](-[#6])-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#7])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6](-[#6])-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6@@H](-[#6])-[#8])-[#6](-[#8])=O |r| |
|---|
| Structure | 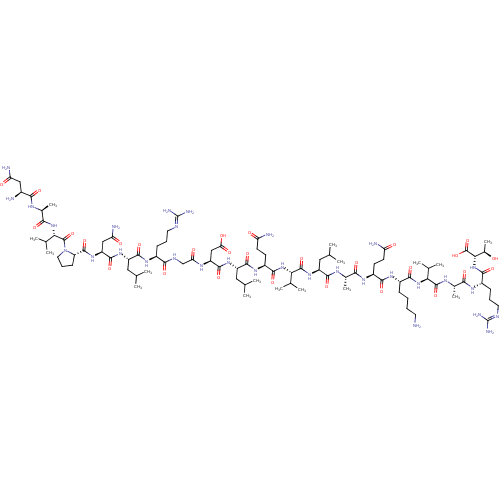
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 DiCara, D; Rapisarda, C; Sutcliffe, JL; Violette, SM; Weinreb, PH; Hart, IR; Howard, MJ; Marshall, JF Structure-function analysis of Arg-Gly-Asp helix motifs in alpha v beta 6 integrin ligands. J Biol Chem282:9657-65 (2007) [PubMed] Article
DiCara, D; Rapisarda, C; Sutcliffe, JL; Violette, SM; Weinreb, PH; Hart, IR; Howard, MJ; Marshall, JF Structure-function analysis of Arg-Gly-Asp helix motifs in alpha v beta 6 integrin ligands. J Biol Chem282:9657-65 (2007) [PubMed] Article