| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 |
|---|
| Ligand | BDBM23078 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_557657 (CHEMBL961607) |
|---|
| pH | 9±n/a |
|---|
| Ki | 0.75±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Slaymaker, IM; Bracey, M; Mileni, M; Garfunkle, J; Cravatt, BF; Boger, DL; Stevens, RC Correlation of inhibitor effects on enzyme activity and thermal stability for the integral membrane protein fatty acid amide hydrolase. Bioorg Med Chem Lett18:5847-50 (2008) [PubMed] Article Slaymaker, IM; Bracey, M; Mileni, M; Garfunkle, J; Cravatt, BF; Boger, DL; Stevens, RC Correlation of inhibitor effects on enzyme activity and thermal stability for the integral membrane protein fatty acid amide hydrolase. Bioorg Med Chem Lett18:5847-50 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 |
|---|
| Name: | Fatty-acid amide hydrolase 1 |
|---|
| Synonyms: | Anandamide amidohydrolase | Anandamide amidohydrolase 1 | FAAH | FAAH1 | FAAH1_HUMAN | Fatty Acid Amide Hydrolase (FAAH) | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Oleamide hydrolase 1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 63071.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O00519 |
|---|
| Residue: | 579 |
|---|
| Sequence: | MVQYELWAALPGASGVALACCFVAAAVALRWSGRRTARGAVVRARQRQRAGLENMDRAAQ
RFRLQNPDLDSEALLALPLPQLVQKLHSRELAPEAVLFTYVGKAWEVNKGTNCVTSYLAD
CETQLSQAPRQGLLYGVPVSLKECFTYKGQDSTLGLSLNEGVPAECDSVVVHVLKLQGAV
PFVHTNVPQSMFSYDCSNPLFGQTVNPWKSSKSPGGSSGGEGALIGSGGSPLGLGTDIGG
SIRFPSSFCGICGLKPTGNRLSKSGLKGCVYGQEAVRLSVGPMARDVESLALCLRALLCE
DMFRLDPTVPPLPFREEVYTSSQPLRVGYYETDNYTMPSPAMRRAVLETKQSLEAAGHTL
VPFLPSNIPHALETLSTGGLFSDGGHTFLQNFKGDFVDPCLGDLVSILKLPQWLKGLLAF
LVKPLLPRLSAFLSNMKSRSAGKLWELQHEIEVYRKTVIAQWRALDLDVVLTPMLAPALD
LNAPGRATGAVSYTMLYNCLDFPAGVVPVTTVTAEDEAQMEHYRGYFGDIWDKMLQKGMK
KSVGLPVAVQCVALPWQEELCLRFMREVERLMTPEKQSS
|
|
|
|---|
| BDBM23078 |
|---|
| n/a |
|---|
| Name | BDBM23078 |
|---|
| Synonyms: | 3-(4-phenylphenyl)-1-[5-(pyridin-2-yl)-1,3-oxazol-2-yl]propan-1-one | CHEMBL227046 | alpha-ketooxazole, 11j |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H18N2O2 |
|---|
| Mol. Mass. | 354.4012 |
|---|
| SMILES | O=C(CCc1ccc(cc1)-c1ccccc1)c1ncc(o1)-c1ccccn1 |
|---|
| Structure | 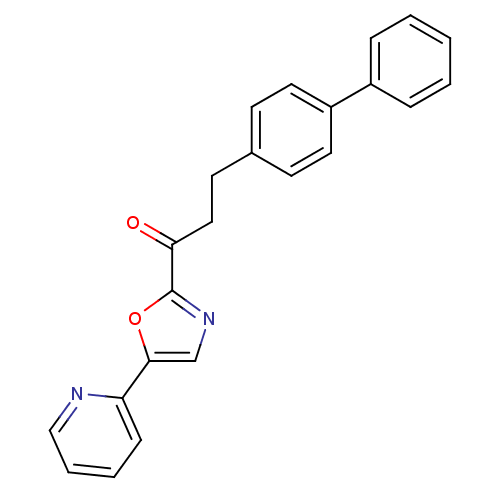
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Slaymaker, IM; Bracey, M; Mileni, M; Garfunkle, J; Cravatt, BF; Boger, DL; Stevens, RC Correlation of inhibitor effects on enzyme activity and thermal stability for the integral membrane protein fatty acid amide hydrolase. Bioorg Med Chem Lett18:5847-50 (2008) [PubMed] Article
Slaymaker, IM; Bracey, M; Mileni, M; Garfunkle, J; Cravatt, BF; Boger, DL; Stevens, RC Correlation of inhibitor effects on enzyme activity and thermal stability for the integral membrane protein fatty acid amide hydrolase. Bioorg Med Chem Lett18:5847-50 (2008) [PubMed] Article