| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Ras guanyl-releasing protein 3 |
|---|
| Ligand | BDBM50244866 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_512396 (CHEMBL979907) |
|---|
| Ki | 1.8±n/a nM |
|---|
| Citation |  Duan, D; Sigano, DM; Kelley, JA; Lai, CC; Lewin, NE; Kedei, N; Peach, ML; Lee, J; Abeyweera, TP; Rotenberg, SA; Kim, H; Kim, YH; El Kazzouli, S; Chung, JU; Young, HA; Young, MR; Baker, A; Colburn, NH; Haimovitz-Friedman, A; Truman, JP; Parrish, DA; Deschamps, JR; Perry, NA; Surawski, RJ; Blumberg, PM; Marquez, VE Conformationally constrained analogues of diacylglycerol. 29. Cells sort diacylglycerol-lactone chemical zip codes to produce diverse and selective biological activities. J Med Chem51:5198-220 (2008) [PubMed] Article Duan, D; Sigano, DM; Kelley, JA; Lai, CC; Lewin, NE; Kedei, N; Peach, ML; Lee, J; Abeyweera, TP; Rotenberg, SA; Kim, H; Kim, YH; El Kazzouli, S; Chung, JU; Young, HA; Young, MR; Baker, A; Colburn, NH; Haimovitz-Friedman, A; Truman, JP; Parrish, DA; Deschamps, JR; Perry, NA; Surawski, RJ; Blumberg, PM; Marquez, VE Conformationally constrained analogues of diacylglycerol. 29. Cells sort diacylglycerol-lactone chemical zip codes to produce diverse and selective biological activities. J Med Chem51:5198-220 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Ras guanyl-releasing protein 3 |
|---|
| Name: | Ras guanyl-releasing protein 3 |
|---|
| Synonyms: | GRP3 | GRP3_HUMAN | KIAA0846 | RAS guanyl releasing protein 3 | RASGRP3 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 78335.20 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1364407 |
|---|
| Residue: | 690 |
|---|
| Sequence: | MGSSGLGKAATLDELLCTCIEMFDDNGELDNSYLPRIVLLMHRWYLSSTELAEKLLCMYR
NATGESCNEFRLKICYFMRYWILKFPAEFNLDLGLIRMTEEFREVASQLGYEKHVSLIDI
SSIPSYDWMRRVTQRKKVSKKGKACLLFDHLEPIELAEHLTFLEHKSFRRISFTDYQSYV
IHGCLENNPTLERSIALFNGISKWVQLMVLSKPTPQQRAEVITKFINVAKKLLQLKNFNT
LMAVVGGLSHSSISRLKETHSHLSSEVTKNWNEMTELVSSNGNYCNYRKAFADCDGFKIP
ILGVHLKDLIAVHVIFPDWTEENKVNIVKMHQLSVTLSELVSLQNASHHLEPNMDLINLL
TLSLDLYHTEDDIYKLSLVLEPRNSKSQPTSPTTPNKPVVPLEWALGVMPKPDPTVINKH
IRKLVESVFRNYDHDHDGYISQEDFESIAANFPFLDSFCVLDKDQDGLISKDEMMAYFLR
AKSQLHCKMGPGFIHNFQEMTYLKPTFCEHCAGFLWGIIKQGYKCKDCGANCHKQCKDLL
VLACRRFARAPSLSSGHGSLPGSPSLPPAQDEVFEFPGVTAGHRDLDSRAITLVTGSSRK
ISVRLQRATTSQATQTEPVWSEAGWGDSGSHTFPKMKSKFHDKAAKDKGFAKWENEKPRV
HAGVDVVDRGTEFELDQDEGEETRQDGEDG
|
|
|
|---|
| BDBM50244866 |
|---|
| n/a |
|---|
| Name | BDBM50244866 |
|---|
| Synonyms: | CHEMBL511820 | rac-(E)-{2-(Hydroxymethyl)-4-[(4-nitrophenyl)methylene]-5-oxo-2-2,3-dihydrofuryl}methyl 2-(Dimethylamino)benzoate |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H22N2O7 |
|---|
| Mol. Mass. | 426.4193 |
|---|
| SMILES | CN(C)c1ccccc1C(=O)OCC1(CO)C\C(=C/c2ccc(cc2)[N+]([O-])=O)C(=O)O1 |
|---|
| Structure | 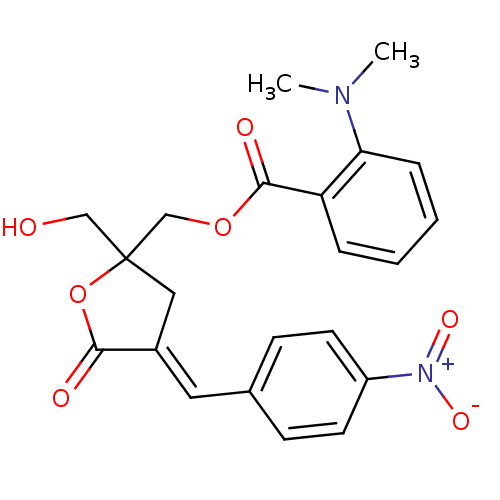
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Duan, D; Sigano, DM; Kelley, JA; Lai, CC; Lewin, NE; Kedei, N; Peach, ML; Lee, J; Abeyweera, TP; Rotenberg, SA; Kim, H; Kim, YH; El Kazzouli, S; Chung, JU; Young, HA; Young, MR; Baker, A; Colburn, NH; Haimovitz-Friedman, A; Truman, JP; Parrish, DA; Deschamps, JR; Perry, NA; Surawski, RJ; Blumberg, PM; Marquez, VE Conformationally constrained analogues of diacylglycerol. 29. Cells sort diacylglycerol-lactone chemical zip codes to produce diverse and selective biological activities. J Med Chem51:5198-220 (2008) [PubMed] Article
Duan, D; Sigano, DM; Kelley, JA; Lai, CC; Lewin, NE; Kedei, N; Peach, ML; Lee, J; Abeyweera, TP; Rotenberg, SA; Kim, H; Kim, YH; El Kazzouli, S; Chung, JU; Young, HA; Young, MR; Baker, A; Colburn, NH; Haimovitz-Friedman, A; Truman, JP; Parrish, DA; Deschamps, JR; Perry, NA; Surawski, RJ; Blumberg, PM; Marquez, VE Conformationally constrained analogues of diacylglycerol. 29. Cells sort diacylglycerol-lactone chemical zip codes to produce diverse and selective biological activities. J Med Chem51:5198-220 (2008) [PubMed] Article