| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 |
|---|
| Ligand | BDBM50240416 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_557788 (CHEMBL953297) |
|---|
| IC50 | 540±n/a nM |
|---|
| Citation |  Nomura, DK; Hudak, CS; Ward, AM; Burston, JJ; Issa, RS; Fisher, KJ; Abood, ME; Wiley, JL; Lichtman, AH; Casida, JE Monoacylglycerol lipase regulates 2-arachidonoylglycerol action and arachidonic acid levels. Bioorg Med Chem Lett18:5875-8 (2008) [PubMed] Article Nomura, DK; Hudak, CS; Ward, AM; Burston, JJ; Issa, RS; Fisher, KJ; Abood, ME; Wiley, JL; Lichtman, AH; Casida, JE Monoacylglycerol lipase regulates 2-arachidonoylglycerol action and arachidonic acid levels. Bioorg Med Chem Lett18:5875-8 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 |
|---|
| Name: | Fatty-acid amide hydrolase 1 |
|---|
| Synonyms: | Anandamide amidohydrolase | Anandamide amidohydrolase 1 | FAAH1_MOUSE | Faah | Faah1 | Fatty Acid Amide Hydrolase | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Oleamide hydrolase 1 |
|---|
| Type: | Hydrolase; single-pass membrane protein; homodimer |
|---|
| Mol. Mass.: | 63227.28 |
|---|
| Organism: | Mus musculus (mouse) |
|---|
| Description: | Mouse brain membranes were used in the assay. |
|---|
| Residue: | 579 |
|---|
| Sequence: | MVLSEVWTALSGLSGVCLACSLLSAAVVLRWTRSQTARGAVTRARQKQRAGLETMDKAVQ
RFRLQNPDLDSEALLALPLLQLVQKLQSGELSPEAVLFTYLGKAWEVNKGTNCVTSYLTD
CETQLSQAPRQGLLYGVPVSLKECFSYKGHASTLGLSLNEGVTSESDCVVVQVLKLQGAV
PFVHTNVPQSMLSYDCSNPLFGQTMNPWKPSKSPGGSSGGEGALIGSGGSPLGLGTDIGG
SIRFPSAFCGICGLKPTGNRLSKSGLKSCVYGQTAVQLSVGPMARDVDSLALCMKALLCE
DLFRLDSTIPPLPFREEIYRSSRPLRVGYYETDNYTMPTPAMRRAVMETKQSLEAAGHTL
VPFLPNNIPYALEVLSAGGLFSDGGCSFLQNFKGDFVDPCLGDLVLVLKLPRWFKKLLSF
LLKPLFPRLAAFLNSMCPRSAEKLWELQHEIEMYRQSVIAQWKAMNLDVVLTPMLGPALD
LNTPGRATGAISYTVLYNCLDFPAGVVPVTTVTAEDDAQMEHYKGYFGDMWDNILKKGMK
KGIGLPVAVQCVALPWQEELCLRFMREVERLMTPEKRPS
|
|
|
|---|
| BDBM50240416 |
|---|
| n/a |
|---|
| Name | BDBM50240416 |
|---|
| Synonyms: | CHEMBL23838 | O,O-diethyl O-p-nitrophenyl phosphate | diethyl 4-nitrophenyl phosphate | diethyl p-nitrophenyl phosphate | diethyl paraoxon | ethyl paraoxon | p-nitrophenyl diethyl phosphate | paraoxon | phosphacol | phosphoric acid diethyl 4-nitrophenyl ester |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H14NO6P |
|---|
| Mol. Mass. | 275.195 |
|---|
| SMILES | CCOP(=O)(OCC)Oc1ccc(cc1)[N+]([O-])=O |
|---|
| Structure | 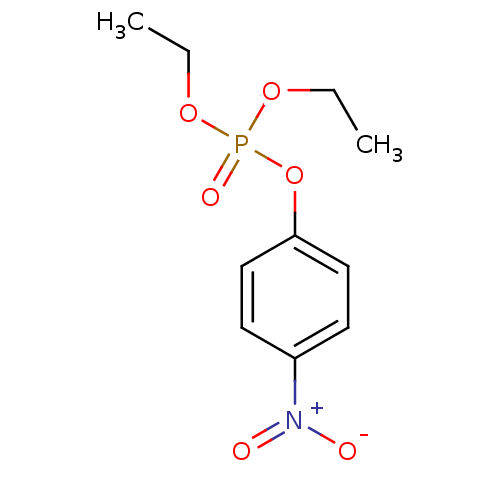
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Nomura, DK; Hudak, CS; Ward, AM; Burston, JJ; Issa, RS; Fisher, KJ; Abood, ME; Wiley, JL; Lichtman, AH; Casida, JE Monoacylglycerol lipase regulates 2-arachidonoylglycerol action and arachidonic acid levels. Bioorg Med Chem Lett18:5875-8 (2008) [PubMed] Article
Nomura, DK; Hudak, CS; Ward, AM; Burston, JJ; Issa, RS; Fisher, KJ; Abood, ME; Wiley, JL; Lichtman, AH; Casida, JE Monoacylglycerol lipase regulates 2-arachidonoylglycerol action and arachidonic acid levels. Bioorg Med Chem Lett18:5875-8 (2008) [PubMed] Article