| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lipoprotein lipase |
|---|
| Ligand | BDBM50254341 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_559993 (CHEMBL1018743) |
|---|
| IC50 | 320±n/a nM |
|---|
| Citation |  Goodman, KB; Bury, MJ; Cheung, M; Cichy-Knight, MA; Dowdell, SE; Dunn, AK; Lee, D; Lieby, JA; Moore, ML; Scherzer, DA; Sha, D; Suarez, DP; Murphy, DJ; Harpel, MR; Manas, ES; McNulty, DE; Annan, RS; Matico, RE; Schwartz, BK; Trill, JJ; Sweitzer, TD; Wang, DY; Keller, PM; Krawiec, JA; Jaye, MC Discovery of potent, selective sulfonylfuran urea endothelial lipase inhibitors. Bioorg Med Chem Lett19:27-30 (2008) [PubMed] Article Goodman, KB; Bury, MJ; Cheung, M; Cichy-Knight, MA; Dowdell, SE; Dunn, AK; Lee, D; Lieby, JA; Moore, ML; Scherzer, DA; Sha, D; Suarez, DP; Murphy, DJ; Harpel, MR; Manas, ES; McNulty, DE; Annan, RS; Matico, RE; Schwartz, BK; Trill, JJ; Sweitzer, TD; Wang, DY; Keller, PM; Krawiec, JA; Jaye, MC Discovery of potent, selective sulfonylfuran urea endothelial lipase inhibitors. Bioorg Med Chem Lett19:27-30 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lipoprotein lipase |
|---|
| Name: | Lipoprotein lipase |
|---|
| Synonyms: | LIPL_RAT | Lpl |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 53093.31 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_559993 |
|---|
| Residue: | 474 |
|---|
| Sequence: | MESKALLLVALGVWLQSLTAFRGGVAAADGGRDFSDIESKFALRTPEDTAEDTCHLIPGL
ADSVSNCHFNHSSKTFVVIHGWTVTGMYESWVPKLVAALYKREPDSNVIVVDWLYRAQQH
YPVSAGYTKLVGNDVARFINWLEEEFNYPLDNVHLLGYSLGAHAAGVAGSLTNKKVNRIT
GLDPAGPNFEYAEAPSRLSPDDADFVDVLHTFTRGSPGRSIGIQKPVGHVDIYPNGGTFQ
PGCNIGEAIRVIAEKGLGDVDQLVKCSHERSIHLFIDSLLNEENPSKAYRCNSKEAFEKG
LCLSCRKNRCNNVGYEINKVRAKRSSKMYLKTRSQMPYKVFHYQVKIHFSGTENDKQNNQ
AFEISLYGTVAESENIPFTLPEVATNKTYSFLIYTEVDIGELLMMKLKWKNDSYFRWSDW
WSSPSFVIEKIRVKAGETQKKVIFCAREKVSHLQKGKDAAVFVKCHDKSLKKSG
|
|
|
|---|
| BDBM50254341 |
|---|
| n/a |
|---|
| Name | BDBM50254341 |
|---|
| Synonyms: | 1-(2-chlorophenyl)-3-(2-methyl-5-(piperidin-1-ylsulfonyl)furan-3-yl)urea | CHEMBL451911 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H20ClN3O4S |
|---|
| Mol. Mass. | 397.876 |
|---|
| SMILES | Cc1oc(cc1NC(=O)Nc1ccccc1Cl)S(=O)(=O)N1CCCCC1 |
|---|
| Structure | 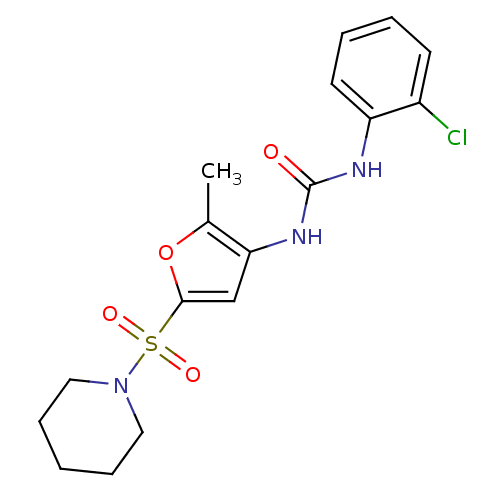
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Goodman, KB; Bury, MJ; Cheung, M; Cichy-Knight, MA; Dowdell, SE; Dunn, AK; Lee, D; Lieby, JA; Moore, ML; Scherzer, DA; Sha, D; Suarez, DP; Murphy, DJ; Harpel, MR; Manas, ES; McNulty, DE; Annan, RS; Matico, RE; Schwartz, BK; Trill, JJ; Sweitzer, TD; Wang, DY; Keller, PM; Krawiec, JA; Jaye, MC Discovery of potent, selective sulfonylfuran urea endothelial lipase inhibitors. Bioorg Med Chem Lett19:27-30 (2008) [PubMed] Article
Goodman, KB; Bury, MJ; Cheung, M; Cichy-Knight, MA; Dowdell, SE; Dunn, AK; Lee, D; Lieby, JA; Moore, ML; Scherzer, DA; Sha, D; Suarez, DP; Murphy, DJ; Harpel, MR; Manas, ES; McNulty, DE; Annan, RS; Matico, RE; Schwartz, BK; Trill, JJ; Sweitzer, TD; Wang, DY; Keller, PM; Krawiec, JA; Jaye, MC Discovery of potent, selective sulfonylfuran urea endothelial lipase inhibitors. Bioorg Med Chem Lett19:27-30 (2008) [PubMed] Article