

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Poly [ADP-ribose] polymerase 1 | ||
| Ligand | BDBM50165496 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_515852 (CHEMBL993111) | ||
| IC50 | 3.8±n/a nM | ||
| Citation |  Zhang, WT; Ruan, JL; Wu, PF; Jiang, FC; Zhang, LN; Fang, W; Chen, XL; Wang, Y; Cao, BS; Chen, GY; Zhu, YJ; Gu, J; Chen, JG Design, synthesis, and cytoprotective effect of 2-aminothiazole analogues as potent poly(ADP-ribose) polymerase-1 inhibitors. J Med Chem52:718-25 (2009) [PubMed] Article Zhang, WT; Ruan, JL; Wu, PF; Jiang, FC; Zhang, LN; Fang, W; Chen, XL; Wang, Y; Cao, BS; Chen, GY; Zhu, YJ; Gu, J; Chen, JG Design, synthesis, and cytoprotective effect of 2-aminothiazole analogues as potent poly(ADP-ribose) polymerase-1 inhibitors. J Med Chem52:718-25 (2009) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Poly [ADP-ribose] polymerase 1 | |||
| Name: | Poly [ADP-ribose] polymerase 1 | ||
| Synonyms: | (ARTD1 or PARP1) | 2.4.2.- | 2.4.2.30 | ADP-ribosyltransferase diphtheria toxin-like 1 | ADPRT | ADPRT 1 | ARTD1 | DNA ADP-ribosyltransferase PARP1 | Human diphtheria toxin-like ADP-ribosyltransferase (ARTD1 or PARP1) | NAD(+) ADP-ribosyltransferase 1 | NT-PARP-1 | PARP-1 | PARP1 | PARP1_HUMAN | PPOL | Poly [ADP-ribose] polymerase (PARP) | Poly [ADP-ribose] polymerase 1 (PARP) | Poly [ADP-ribose] polymerase 1 (PARP-1) | Poly [ADP-ribose] polymerase 1 (PARP1) | Poly [ADP-ribose] polymerase 1, 24-kDa form | Poly [ADP-ribose] polymerase 1, 28-kDa form | Poly [ADP-ribose] polymerase 1, 89-kDa form | Poly [ADP-ribose] polymerase 1, processed C-terminus | Poly [ADP-ribose] polymerase 1, processed N-terminus | Poly [ADP-ribose] polymerase-1 | Poly(ADP-ribose) polymerase 1 (PARP1) | Poly(ADP-ribose) polymerase-1 (ARTD1/PARP1) | Poly[ADP-ribose] synthase 1 | Protein poly-ADP-ribosyltransferase PARP1 | Synonyms=ADPRT | ||
| Type: | n/a | ||
| Mol. Mass.: | 113114.22 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P09874 | ||
| Residue: | 1014 | ||
| Sequence: |
| ||
| BDBM50165496 | |||
| n/a | |||
| Name | BDBM50165496 | ||
| Synonyms: | 1-(2-fluoro-5-((4-oxo-3,4-dihydrophthalazin-1-yl)methyl)phenyl)-3-methylpyrrolidine-2,5-dione | 1-[2-Fluoro-5-(4-oxo-3,4-dihydro-phthalazin-1-ylmethyl)-phenyl]-3-methyl-pyrrolidine-2,5-dione | CHEMBL370838 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H16FN3O3 | ||
| Mol. Mass. | 365.3577 | ||
| SMILES | Cc1cc(O)n(c1O)-c1cc(Cc2n[nH]c(=O)c3ccccc23)ccc1F |(2.63,-45.05,;1.1,-44.9,;.32,-43.57,;-1.18,-43.9,;-2.34,-42.88,;-1.33,-45.44,;.08,-46.05,;.41,-47.56,;-2.66,-46.22,;-3.99,-45.46,;-5.31,-46.24,;-6.65,-45.47,;-6.65,-43.93,;-5.32,-43.16,;-5.32,-41.61,;-6.67,-40.84,;-6.68,-39.3,;-8,-41.62,;-9.34,-40.86,;-10.66,-41.63,;-10.67,-43.18,;-9.33,-43.95,;-7.99,-43.17,;-5.31,-47.78,;-3.98,-48.54,;-2.65,-47.77,;-1.31,-48.53,)| | ||
| Structure | 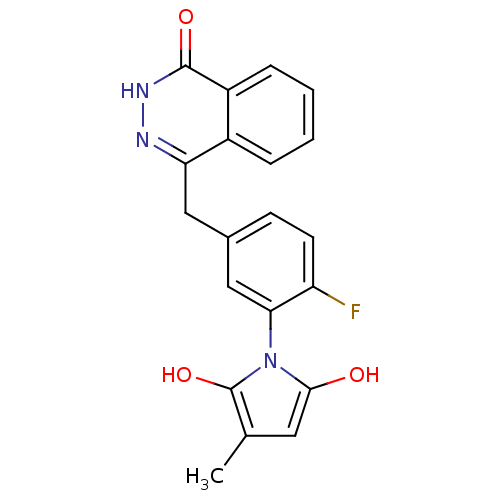
| ||