| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Protein mono-ADP-ribosyltransferase PARP3 [R100H] |
|---|
| Ligand | BDBM50226518 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_501487 (CHEMBL984126) |
|---|
| Kd | >2000±n/a nM |
|---|
| Citation |  Lehtiö, L; Jemth, AS; Collins, R; Loseva, O; Johansson, A; Markova, N; Hammarström, M; Flores, A; Holmberg-Schiavone, L; Weigelt, J; Helleday, T; Schüler, H; Karlberg, T Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3. J Med Chem52:3108-11 (2009) [PubMed] Article Lehtiö, L; Jemth, AS; Collins, R; Loseva, O; Johansson, A; Markova, N; Hammarström, M; Flores, A; Holmberg-Schiavone, L; Weigelt, J; Helleday, T; Schüler, H; Karlberg, T Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3. J Med Chem52:3108-11 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Protein mono-ADP-ribosyltransferase PARP3 [R100H] |
|---|
| Name: | Protein mono-ADP-ribosyltransferase PARP3 [R100H] |
|---|
| Synonyms: | (ARTD3 or PARP3) | ADP-ribosyltransferase diphtheria toxin-like 3 | ADPRT3 | ADPRTL3 | Human diphtheria toxin-like ADP-ribosyltransferase (ARTD3 or PARP3) | PARP3 | PARP3_HUMAN | Poly [ADP-ribose] polymerase 3 | Poly [ADP-ribose] polymerase 3 (PARP-3) | Poly(ADP-ribose) polymerase-3 (ARTD3/PARP3) |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 60071.85 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9Y6F1[R100H] |
|---|
| Residue: | 533 |
|---|
| Sequence: | MAPKPKPWVQTEGPEKKKGRQAGREEDPFRSTAEALKAIPAEKRIIRVDPTCPLSSNPGT
QVYEDYNCTLNQTNIENNNNKFYIIQLLQDSNRFFTCWNHWGRVGEVGQSKINHFTRLED
AKKDFEKKFREKTKNNWAERDHFVSHPGKYTLIEVQAEDEAQEAVVKVDRGPVRTVTKRV
QPCSLDPATQKLITNIFSKEMFKNTMALMDLDVKKMPLGKLSKQQIARGFEALEALEEAL
KGPTDGGQSLEELSSHFYTVIPHNFGHSQPPPINSPELLQAKKDMLLVLADIELAQALQA
VSEQEKTVEEVPHPLDRDYQLLKCQLQLLDSGAPEYKVIQTYLEQTGSNHRCPTLQHIWK
VNQEGEEDRFQAHSKLGNRKLLWHGTNMAVVAAILTSGLRIMPHSGGRVGKGIYFASENS
KSAGYVIGMKCGAHHVGYMFLGEVALGREHHINTDNPSLKSPPPGFDSVIARGHTEPDPT
QDTELELDGQQVVVPQGQPVPCPEFSSSTFSQSEYLIYQESQCRLRYLLEVHL
|
|
|
|---|
| BDBM50226518 |
|---|
| n/a |
|---|
| Name | BDBM50226518 |
|---|
| Synonyms: | 3-Aminobenzoesaeure | 3-aminobenzoic acid | CHEMBL307782 | m-Aminobenzoesaeure |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C7H7NO2 |
|---|
| Mol. Mass. | 137.136 |
|---|
| SMILES | Nc1cccc(c1)C(O)=O |
|---|
| Structure | 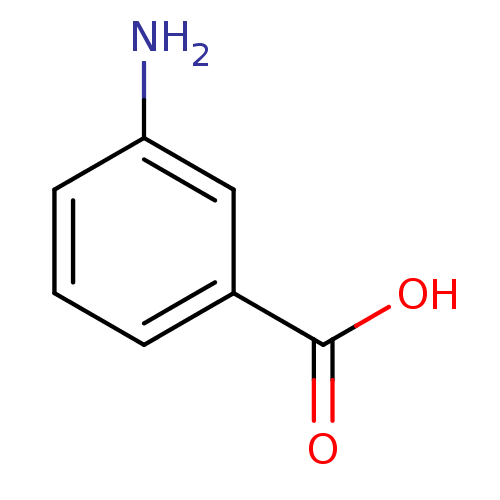
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lehtiö, L; Jemth, AS; Collins, R; Loseva, O; Johansson, A; Markova, N; Hammarström, M; Flores, A; Holmberg-Schiavone, L; Weigelt, J; Helleday, T; Schüler, H; Karlberg, T Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3. J Med Chem52:3108-11 (2009) [PubMed] Article
Lehtiö, L; Jemth, AS; Collins, R; Loseva, O; Johansson, A; Markova, N; Hammarström, M; Flores, A; Holmberg-Schiavone, L; Weigelt, J; Helleday, T; Schüler, H; Karlberg, T Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3. J Med Chem52:3108-11 (2009) [PubMed] Article