| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine/threonine-protein kinase pim-1 |
|---|
| Ligand | BDBM26697 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_500013 (CHEMBL1022925) |
|---|
| Ki | 22±n/a nM |
|---|
| Citation |  Grey, R; Pierce, AC; Bemis, GW; Jacobs, MD; Moody, CS; Jajoo, R; Mohal, N; Green, J Structure-based design of 3-aryl-6-amino-triazolo[4,3-b]pyridazine inhibitors of Pim-1 kinase. Bioorg Med Chem Lett19:3019-22 (2009) [PubMed] Article Grey, R; Pierce, AC; Bemis, GW; Jacobs, MD; Moody, CS; Jajoo, R; Mohal, N; Green, J Structure-based design of 3-aryl-6-amino-triazolo[4,3-b]pyridazine inhibitors of Pim-1 kinase. Bioorg Med Chem Lett19:3019-22 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Serine/threonine-protein kinase pim-1 |
|---|
| Name: | Serine/threonine-protein kinase pim-1 |
|---|
| Synonyms: | PIM-1 Kinase | PIM1 | PIM1_HUMAN | Proto-oncogene serine/threonine-protein kinase Pim-1 | Serine/threonine-protein kinase (PIM1) | Serine/threonine-protein kinase PIM | Serine/threonine-protein kinase PIM1 | Serine/threonine-protein kinase pim-1 (PIM1) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 35681.82 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P11309 |
|---|
| Residue: | 313 |
|---|
| Sequence: | MLLSKINSLAHLRAAPCNDLHATKLAPGKEKEPLESQYQVGPLLGSGGFGSVYSGIRVSD
NLPVAIKHVEKDRISDWGELPNGTRVPMEVVLLKKVSSGFSGVIRLLDWFERPDSFVLIL
ERPEPVQDLFDFITERGALQEELARSFFWQVLEAVRHCHNCGVLHRDIKDENILIDLNRG
ELKLIDFGSGALLKDTVYTDFDGTRVYSPPEWIRYHRYHGRSAAVWSLGILLYDMVCGDI
PFEHDEEIIRGQVFFRQRVSSECQHLIRWCLALRPSDRPTFEEIQNHPWMQDVLLPQETA
EIHLHSLSPGPSK
|
|
|
|---|
| BDBM26697 |
|---|
| n/a |
|---|
| Name | BDBM26697 |
|---|
| Synonyms: | 7-chloro-9-ethyl-6-fluoro-1H,3H,4H,9H-quinolino[2,3-c][1,2]oxazole-3,4-dione | CHEMBL468280 | Isoxazoloquinolinedione, 1 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C12H8ClFN2O3 |
|---|
| Mol. Mass. | 282.655 |
|---|
| SMILES | CCn1c2noc(=O)c2c(O)c2cc(F)c(Cl)cc12 |
|---|
| Structure | 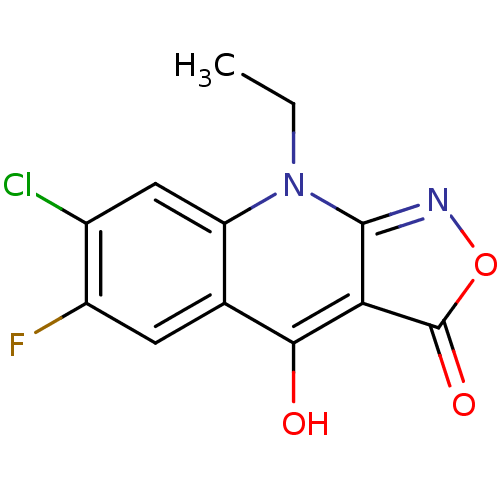
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Grey, R; Pierce, AC; Bemis, GW; Jacobs, MD; Moody, CS; Jajoo, R; Mohal, N; Green, J Structure-based design of 3-aryl-6-amino-triazolo[4,3-b]pyridazine inhibitors of Pim-1 kinase. Bioorg Med Chem Lett19:3019-22 (2009) [PubMed] Article
Grey, R; Pierce, AC; Bemis, GW; Jacobs, MD; Moody, CS; Jajoo, R; Mohal, N; Green, J Structure-based design of 3-aryl-6-amino-triazolo[4,3-b]pyridazine inhibitors of Pim-1 kinase. Bioorg Med Chem Lett19:3019-22 (2009) [PubMed] Article