

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Hepatocyte growth factor receptor | ||
| Ligand | BDBM50258424 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_500028 (CHEMBL1025476) | ||
| Ki | >4000±n/a nM | ||
| Citation |  Grey, R; Pierce, AC; Bemis, GW; Jacobs, MD; Moody, CS; Jajoo, R; Mohal, N; Green, J Structure-based design of 3-aryl-6-amino-triazolo[4,3-b]pyridazine inhibitors of Pim-1 kinase. Bioorg Med Chem Lett19:3019-22 (2009) [PubMed] Article Grey, R; Pierce, AC; Bemis, GW; Jacobs, MD; Moody, CS; Jajoo, R; Mohal, N; Green, J Structure-based design of 3-aryl-6-amino-triazolo[4,3-b]pyridazine inhibitors of Pim-1 kinase. Bioorg Med Chem Lett19:3019-22 (2009) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Hepatocyte growth factor receptor | |||
| Name: | Hepatocyte growth factor receptor | ||
| Synonyms: | Hepatocyte growth factor receptor | Hepatocyte growth factor receptor (MET) | Hepatocyte growth factor receptor (c-MET) | Hepatocyte growth factor receptor (cMET) | MET | MET_HUMAN | Met proto-oncogene (hepatocyte growth factor receptor) | Proto-oncogene c-Met | Tyrosine-protein kinase Met (c-Met) | Tyrosine-protein kinase Met (cMet) | c-Met kinase | ||
| Type: | Protein | ||
| Mol. Mass.: | 155559.73 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P08581 | ||
| Residue: | 1390 | ||
| Sequence: |
| ||
| BDBM50258424 | |||
| n/a | |||
| Name | BDBM50258424 | ||
| Synonyms: | CHEMBL494221 | trans-4-(3-(3-(trifluoromethyl)phenyl)-[1,2,4]triazolo[4,3-b]pyridazin-6-ylamino)cyclohexanol | ||
| Type | Small organic molecule | ||
| Emp. Form. | C18H18F3N5O | ||
| Mol. Mass. | 377.3636 | ||
| SMILES | O[C@H]1CC[C@@H](CC1)Nc1ccc2nnc(-c3cccc(c3)C(F)(F)F)n2n1 |r,wU:1.0,wD:4.7,(9.27,-14.54,;10.6,-15.32,;11.94,-14.55,;13.26,-15.32,;13.27,-16.87,;11.93,-17.64,;10.6,-16.86,;14.6,-17.63,;15.94,-16.86,;15.94,-15.32,;17.27,-14.54,;18.59,-15.32,;20.06,-14.85,;20.97,-16.09,;20.06,-17.34,;20.82,-18.68,;20.03,-20,;20.78,-21.34,;22.32,-21.36,;23.11,-20.02,;22.35,-18.69,;24.65,-20.03,;26.17,-20.13,;24.7,-18.5,;24.61,-21.57,;18.59,-16.86,;17.27,-17.62,)| | ||
| Structure | 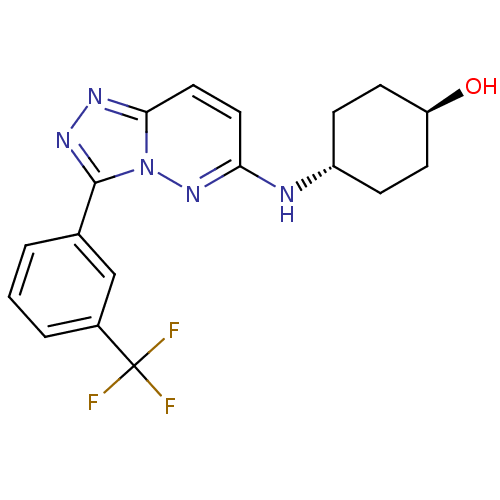
| ||