

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Carbonic anhydrase | ||
| Ligand | BDBM50258299 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_519732 (CHEMBL938995) | ||
| Ki | 0.93±n/a nM | ||
| Citation |  Güzel, O; Maresca, A; Scozzafava, A; Salman, A; Balaban, AT; Supuran, CT Discovery of low nanomolar and subnanomolar inhibitors of the mycobacterial beta-carbonic anhydrases Rv1284 and Rv3273. J Med Chem52:4063-7 (2009) [PubMed] Article Güzel, O; Maresca, A; Scozzafava, A; Salman, A; Balaban, AT; Supuran, CT Discovery of low nanomolar and subnanomolar inhibitors of the mycobacterial beta-carbonic anhydrases Rv1284 and Rv3273. J Med Chem52:4063-7 (2009) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Carbonic anhydrase | |||
| Name: | Carbonic anhydrase | ||
| Synonyms: | PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 80567.22 | ||
| Organism: | Mycobacterium tuberculosis | ||
| Description: | ChEMBL_878775 | ||
| Residue: | 764 | ||
| Sequence: |
| ||
| BDBM50258299 | |||
| n/a | |||
| Name | BDBM50258299 | ||
| Synonyms: | 1-({[5-(Aminosulfonyl)-3-(2,3,4,5,6-pentafluorophenyl)-1H-indol-2-yl]carbonyl}amino)-2,4,6 trimethylpyridinium perchlorate | 2,4,6-trimethyl-1-(3-(perfluorophenyl)-5-sulfamoyl-1H-indole-2-carboxamido)pyridinium perchlorate | CHEMBL501294 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H18F5N4O3S | ||
| Mol. Mass. | 525.47 | ||
| SMILES | Cc1cc(C)[n+](NC(=O)c2[nH]c3ccc(cc3c2-c2c(F)c(F)c(F)c(F)c2F)S(N)(=O)=O)c(C)c1 |(8.2,-44.53,;6.87,-45.29,;6.87,-46.84,;5.53,-47.6,;5.53,-49.14,;4.2,-46.84,;2.87,-47.6,;1.53,-46.83,;1.53,-45.29,;.2,-47.6,;.06,-49.13,;-1.45,-49.47,;-2.22,-50.81,;-3.76,-50.81,;-4.53,-49.48,;-3.76,-48.15,;-2.24,-48.15,;-1.21,-46.98,;-1.64,-45.03,;-.5,-44,;.97,-44.47,;-.83,-42.49,;.31,-41.45,;-2.29,-42.02,;-2.62,-40.52,;-3.43,-43.06,;-4.9,-42.59,;-3.1,-44.57,;-4.24,-45.6,;-6.07,-49.48,;-7.59,-49.48,;-6.07,-47.95,;-6.07,-51.03,;4.2,-45.29,;2.86,-44.53,;5.53,-44.53,)| | ||
| Structure | 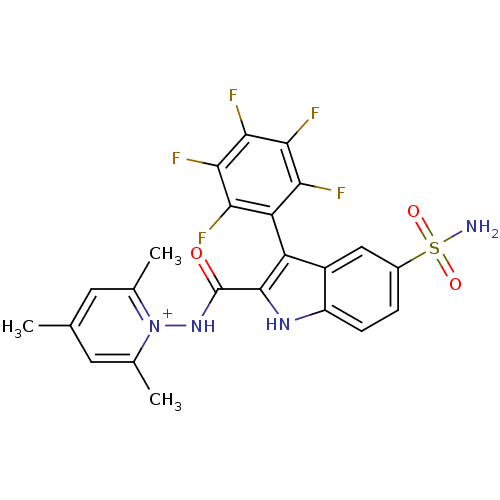
| ||