| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Neuromedin-K receptor |
|---|
| Ligand | BDBM50001450 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_143195 |
|---|
| IC50 | 717±n/a nM |
|---|
| Citation |  Horwell, DC; Morrell, AI; Roberts, E The design and synthesis of non-peptide ligands with affinity and selectivity for tachykinin receptors Bioorg Med Chem Lett6:165-166 (1996) Article Horwell, DC; Morrell, AI; Roberts, E The design and synthesis of non-peptide ligands with affinity and selectivity for tachykinin receptors Bioorg Med Chem Lett6:165-166 (1996) Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Neuromedin-K receptor |
|---|
| Name: | Neuromedin-K receptor |
|---|
| Synonyms: | NK3R_CAVPO | Neurokinin 3 receptor | Neurokinin NK3 | Neuromedin-K receptor | TACR3 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 49456.44 |
|---|
| Organism: | GUINEA PIG |
|---|
| Description: | Neurokinin NK3 TACR3 GUINEA PIG::P30098 |
|---|
| Residue: | 440 |
|---|
| Sequence: | MASPAGNLSAWPGWGWPPPAALRNLTSSPAPTASPSPAPSWTPSPRPGPAHPFLQPPWAV
ALWSLAYGAVVAVAVLGNLVVIWIVLAHKRMRTVTNSFLVNLAFADAAMAALNALVNFIY
ALHGEWYFGANYCRFQNFFPITAVFASIYSMTAIAVDRYMAIIDPLKPRLSATATRIVIG
SIWILAFLLAFPQCLYSKIKVMPGRTLCYVQWPEGSRQHFTYHMIVIVLVYCFPLLIMGI
TYTIVGITLWGGEIPGDTCDKYQEQLKAKRKVVKMMIIVVVTFAICWLPYHIYFILTAIY
QQLNRWKYIQQVYLASFWLAMSSTMYNPIIYCCLNKRFRAGFKRAFRWCPFIHVSSYDEL
ELKATRLHPMRQSSLYTVTRMESMSVVFDSNDGDSARSSHQKRGTTRDVGSNVCSRRNSK
STSTTASFVSSSHMSVEEGS
|
|
|
|---|
| BDBM50001450 |
|---|
| n/a |
|---|
| Name | BDBM50001450 |
|---|
| Synonyms: | (SP)Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2 | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met NH2 | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2 | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2(Substance P) | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2.(Substance P) | Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-amine | ArgProLysProGlnGlnPhePheGlyLeuMet | CHEMBL235363 | H-Arg-Pro-Lys-Pro-Gln-Gln-Phe-D-Phe-Gly-Leu-Met-NH2 | H-Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-Met-NH2(Substance P) | Substance P | Substance P (Arg-Pro-Lys-Pro-Gln-Gln-Phe-Phe-Gly-Leu-MetNH2) | Substance P analogue | tachykinin substance P (SP) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C63H98N18O13S |
|---|
| Mol. Mass. | 1347.63 |
|---|
| SMILES | [#6]-[#16]-[#6]-[#6]-[#6@H](-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#6])-[#6])-[#7]-[#6](=O)-[#6]-[#7]-[#6](=O)-[#6@H](-[#6]-c1ccccc1)-[#7]-[#6](=O)-[#6@H](-[#6]-c1ccccc1)-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@@H]-1-[#6]-[#6]-[#6]-[#7]-1-[#6](=O)-[#6@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#7]-[#6](=O)-[#6@@H]-1-[#6]-[#6]-[#6]-[#7]-1-[#6](=O)-[#6@@H](-[#7])-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6](-[#7])=O |r| |
|---|
| Structure | 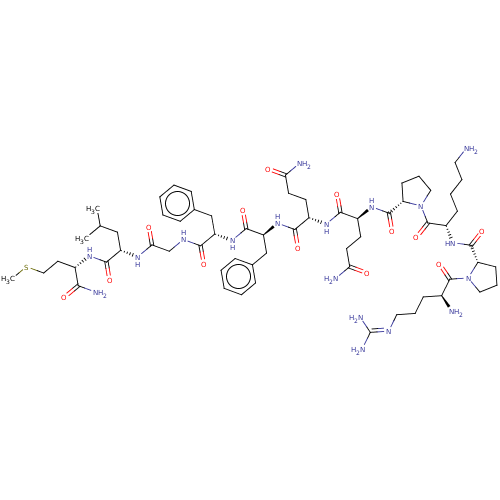
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Horwell, DC; Morrell, AI; Roberts, E The design and synthesis of non-peptide ligands with affinity and selectivity for tachykinin receptors Bioorg Med Chem Lett6:165-166 (1996) Article
Horwell, DC; Morrell, AI; Roberts, E The design and synthesis of non-peptide ligands with affinity and selectivity for tachykinin receptors Bioorg Med Chem Lett6:165-166 (1996) Article