| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Retinoic acid receptor RXR-alpha |
|---|
| Ligand | BDBM50032675 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_589774 (CHEMBL1061278) |
|---|
| Ki | 21±n/a nM |
|---|
| Citation |  Wagner, CE; Jurutka, PW; Marshall, PA; Groy, TL; van der Vaart, A; Ziller, JW; Furmick, JK; Graeber, ME; Matro, E; Miguel, BV; Tran, IT; Kwon, J; Tedeschi, JN; Moosavi, S; Danishyar, A; Philp, JS; Khamees, RO; Jackson, JN; Grupe, DK; Badshah, SL; Hart, JW Modeling, synthesis and biological evaluation of potential retinoid X receptor (RXR) selective agonists: novel analogues of 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)ethynyl]benzoic acid (bexarotene). J Med Chem52:5950-66 (2009) [PubMed] Article Wagner, CE; Jurutka, PW; Marshall, PA; Groy, TL; van der Vaart, A; Ziller, JW; Furmick, JK; Graeber, ME; Matro, E; Miguel, BV; Tran, IT; Kwon, J; Tedeschi, JN; Moosavi, S; Danishyar, A; Philp, JS; Khamees, RO; Jackson, JN; Grupe, DK; Badshah, SL; Hart, JW Modeling, synthesis and biological evaluation of potential retinoid X receptor (RXR) selective agonists: novel analogues of 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)ethynyl]benzoic acid (bexarotene). J Med Chem52:5950-66 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Retinoic acid receptor RXR-alpha |
|---|
| Name: | Retinoic acid receptor RXR-alpha |
|---|
| Synonyms: | NR2B1 | Nuclear receptor subfamily 2 group B member 1 | Nuclear receptor subfamily 4 group A member 2 | Nuclear receptor subfamily 4 group A member 2/Retinoic acid receptor RXR-alpha | RXRA | RXRA_HUMAN | Retinoic acid receptor RXR-alpha/gamma | Retinoid X receptor alpha | Retinoid receptor |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 50820.38 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1456363 |
|---|
| Residue: | 462 |
|---|
| Sequence: | MDTKHFLPLDFSTQVNSSLTSPTGRGSMAAPSLHPSLGPGIGSPGQLHSPISTLSSPING
MGPPFSVISSPMGPHSMSVPTTPTLGFSTGSPQLSSPMNPVSSSEDIKPPLGLNGVLKVP
AHPSGNMASFTKHICAICGDRSSGKHYGVYSCEGCKGFFKRTVRKDLTYTCRDNKDCLID
KRQRNRCQYCRYQKCLAMGMKREAVQEERQRGKDRNENEVESTSSANEDMPVERILEAEL
AVEPKTETYVEANMGLNPSSPNDPVTNICQAADKQLFTLVEWAKRIPHFSELPLDDQVIL
LRAGWNELLIASFSHRSIAVKDGILLATGLHVHRNSAHSAGVGAIFDRVLTELVSKMRDM
QMDKTELGCLRAIVLFNPDSKGLSNPAEVEALREKVYASLEAYCKHKYPEQPGRFAKLLL
RLPALRSIGLKCLEHLFFFKLIGDTPIDTFLMEMLEAPHQMT
|
|
|
|---|
| BDBM50032675 |
|---|
| n/a |
|---|
| Name | BDBM50032675 |
|---|
| Synonyms: | 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)vinyl]benzoic acid | BEXAROTENE | CHEMBL1023 | p-(1-(5,6,7,8-Tetrahydro-3,5,5,8,8-pentamethyl-2-naphthyl)vinyl)benzoic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H28O2 |
|---|
| Mol. Mass. | 348.4779 |
|---|
| SMILES | Cc1cc2c(cc1C(=C)c1ccc(cc1)C(O)=O)C(C)(C)CCC2(C)C |
|---|
| Structure | 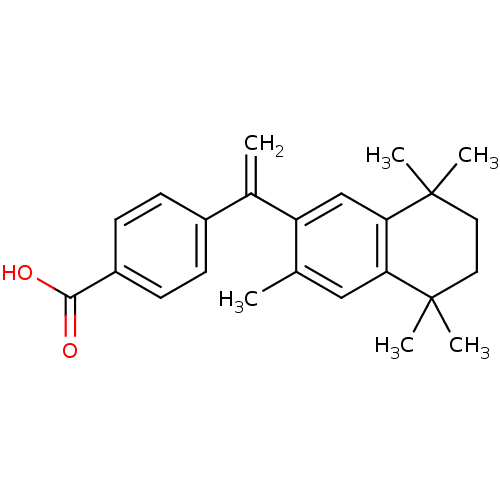
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wagner, CE; Jurutka, PW; Marshall, PA; Groy, TL; van der Vaart, A; Ziller, JW; Furmick, JK; Graeber, ME; Matro, E; Miguel, BV; Tran, IT; Kwon, J; Tedeschi, JN; Moosavi, S; Danishyar, A; Philp, JS; Khamees, RO; Jackson, JN; Grupe, DK; Badshah, SL; Hart, JW Modeling, synthesis and biological evaluation of potential retinoid X receptor (RXR) selective agonists: novel analogues of 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)ethynyl]benzoic acid (bexarotene). J Med Chem52:5950-66 (2009) [PubMed] Article
Wagner, CE; Jurutka, PW; Marshall, PA; Groy, TL; van der Vaart, A; Ziller, JW; Furmick, JK; Graeber, ME; Matro, E; Miguel, BV; Tran, IT; Kwon, J; Tedeschi, JN; Moosavi, S; Danishyar, A; Philp, JS; Khamees, RO; Jackson, JN; Grupe, DK; Badshah, SL; Hart, JW Modeling, synthesis and biological evaluation of potential retinoid X receptor (RXR) selective agonists: novel analogues of 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)ethynyl]benzoic acid (bexarotene). J Med Chem52:5950-66 (2009) [PubMed] Article