| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 3-dehydroquinate dehydratase |
|---|
| Ligand | BDBM50303443 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_595938 (CHEMBL1041787) |
|---|
| Ki | 920±n/a nM |
|---|
| Citation |  Prazeres, VF; Tizón, L; Otero, JM; Guardado-Calvo, P; Llamas-Saiz, AL; van Raaij, MJ; Castedo, L; Lamb, H; Hawkins, AR; González-Bello, C Synthesis and biological evaluation of new nanomolar competitive inhibitors of Helicobacter pylori type II dehydroquinase. Structural details of the role of the aromatic moieties with essential residues. J Med Chem53:191-200 (2010) [PubMed] Article Prazeres, VF; Tizón, L; Otero, JM; Guardado-Calvo, P; Llamas-Saiz, AL; van Raaij, MJ; Castedo, L; Lamb, H; Hawkins, AR; González-Bello, C Synthesis and biological evaluation of new nanomolar competitive inhibitors of Helicobacter pylori type II dehydroquinase. Structural details of the role of the aromatic moieties with essential residues. J Med Chem53:191-200 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 3-dehydroquinate dehydratase |
|---|
| Name: | 3-dehydroquinate dehydratase |
|---|
| Synonyms: | AROQ_HELPY | aroQ |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 18479.08 |
|---|
| Organism: | Helicobacter pylori |
|---|
| Description: | ChEMBL_766424 |
|---|
| Residue: | 167 |
|---|
| Sequence: | MKILVIQGPNLNMLGHRDPRLYGMVTLDQIHEIMQTFVKQGNLDVELEFFQTNFEGEIID
KIQESVGSDYEGIIINPGAFSHTSIAIADAIMLAGKPVIEVHLTNIQAREEFRKNSYTGA
ACGGVIMGFGPLGYNMALMAMVNILAEMKAFQEAQKNNPNNPINNQK
|
|
|
|---|
| BDBM50303443 |
|---|
| n/a |
|---|
| Name | BDBM50303443 |
|---|
| Synonyms: | CHEMBL567136 | Sodium(1R,4S,5R)-1,4-Dihydroxy-3-(thien-3-yl)methoxycyclohex-2-en-1-carboxylate |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C12H13O6S |
|---|
| Mol. Mass. | 285.294 |
|---|
| SMILES | O[C@@H]1C[C@@](O)(C=C(OCc2cccs2)[C@H]1O)C([O-])=O |r,t:5| |
|---|
| Structure | 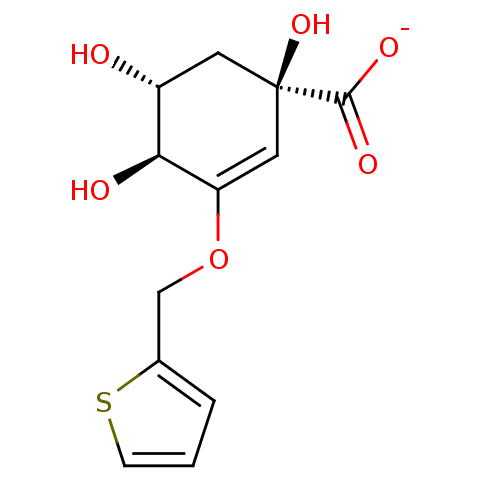
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Prazeres, VF; Tizón, L; Otero, JM; Guardado-Calvo, P; Llamas-Saiz, AL; van Raaij, MJ; Castedo, L; Lamb, H; Hawkins, AR; González-Bello, C Synthesis and biological evaluation of new nanomolar competitive inhibitors of Helicobacter pylori type II dehydroquinase. Structural details of the role of the aromatic moieties with essential residues. J Med Chem53:191-200 (2010) [PubMed] Article
Prazeres, VF; Tizón, L; Otero, JM; Guardado-Calvo, P; Llamas-Saiz, AL; van Raaij, MJ; Castedo, L; Lamb, H; Hawkins, AR; González-Bello, C Synthesis and biological evaluation of new nanomolar competitive inhibitors of Helicobacter pylori type II dehydroquinase. Structural details of the role of the aromatic moieties with essential residues. J Med Chem53:191-200 (2010) [PubMed] Article