

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Delta-type opioid receptor | ||
| Ligand | BDBM50305086 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_603078 (CHEMBL1048561) | ||
| Ki | >10000±n/a nM | ||
| Citation |  Li, W; Tao, YM; Tang, Y; Xu, XJ; Chen, J; Fu, W; Wang, XH; Chao, B; Sheng, W; Xie, Q; Qiu, ZB; Liu, JG Highly selective and potent mu opioid ligands by unexpected substituent on morphine skeleton. Bioorg Med Chem Lett20:418-21 (2010) [PubMed] Article Li, W; Tao, YM; Tang, Y; Xu, XJ; Chen, J; Fu, W; Wang, XH; Chao, B; Sheng, W; Xie, Q; Qiu, ZB; Liu, JG Highly selective and potent mu opioid ligands by unexpected substituent on morphine skeleton. Bioorg Med Chem Lett20:418-21 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Delta-type opioid receptor | |||
| Name: | Delta-type opioid receptor | ||
| Synonyms: | Cytochrome P450 3A4 | DOR-1 | Delta opioid receptor | Delta-type opioid receptor | Delta-type opioid receptor (DOR) | OPIATE Delta | OPRD_RAT | Opiate Delta 1 | Opioid receptor | Opioid receptor A | Opioid receptors; mu & delta | Oprd1 | Ror-a | Voltage-gated potassium channel | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 40465.04 | ||
| Organism: | Rattus norvegicus (rat) | ||
| Description: | Competition binding assays were using CHO-K1 cell membranes expressing the opioid receptor. | ||
| Residue: | 372 | ||
| Sequence: |
| ||
| BDBM50305086 | |||
| n/a | |||
| Name | BDBM50305086 | ||
| Synonyms: | 21-nitro-7alpha-phenyl-6alpha,14alpha-endo-Ethenotetrahydrothebaine | CHEMBL592952 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C27H28N2O5 | ||
| Mol. Mass. | 460.5216 | ||
| SMILES | COc1ccc2C[C@H]3N(C)CC[C@@]45[C@@H](Oc1c24)[C@]1(OC)C=C[C@@]35C[C@@H]1c1cccc(c1)[N+]([O-])=O |r,wU:7.7,17.20,wD:12.18,22.24,24.30,13.13,c:23,(-7.57,-14.62,;-6.24,-13.85,;-4.89,-14.63,;-3.57,-13.86,;-2.23,-14.62,;-2.23,-16.17,;-.89,-16.94,;-.89,-18.49,;.44,-19.26,;1.59,-20.28,;-.7,-17.7,;-2.24,-17.7,;-3.56,-18.48,;-4.9,-19.25,;-5.68,-17.5,;-4.9,-16.17,;-3.56,-16.94,;-4.9,-20.79,;-6.37,-21.24,;-6.72,-22.74,;-3.53,-20.81,;-3.54,-19.25,;-2.23,-19.26,;-2.23,-20.79,;-3.56,-21.57,;-3.56,-23.11,;-4.9,-23.87,;-4.89,-25.41,;-3.56,-26.18,;-2.22,-25.4,;-2.23,-23.86,;-.88,-26.16,;-.88,-27.7,;.45,-25.39,)| | ||
| Structure | 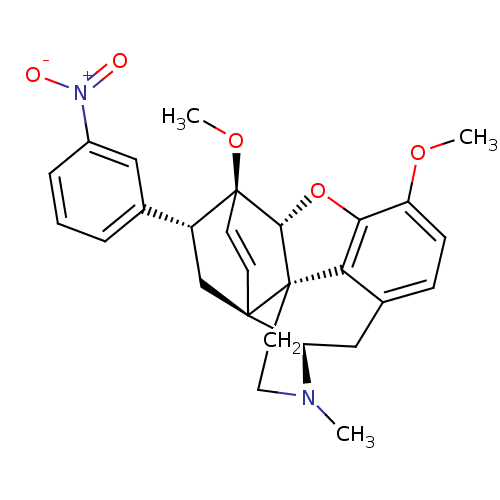
| ||