| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosine-protein kinase ZAP-70 |
|---|
| Ligand | BDBM50305126 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_603256 (CHEMBL1049480) |
|---|
| IC50 | >1000±n/a nM |
|---|
| Citation |  Takayama, T; Umemiya, H; Amada, H; Yabuuchi, T; Koami, T; Shiozawa, F; Oka, Y; Takaoka, A; Yamaguchi, A; Endo, M; Sato, M Ring-fused pyrazole derivatives as potent inhibitors of lymphocyte-specific kinase (Lck): Structure, synthesis, and SAR. Bioorg Med Chem Lett20:112-6 (2010) [PubMed] Article Takayama, T; Umemiya, H; Amada, H; Yabuuchi, T; Koami, T; Shiozawa, F; Oka, Y; Takaoka, A; Yamaguchi, A; Endo, M; Sato, M Ring-fused pyrazole derivatives as potent inhibitors of lymphocyte-specific kinase (Lck): Structure, synthesis, and SAR. Bioorg Med Chem Lett20:112-6 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Tyrosine-protein kinase ZAP-70 |
|---|
| Name: | Tyrosine-protein kinase ZAP-70 |
|---|
| Synonyms: | 70 kDa zeta-associated protein | SRK | Syk-related tyrosine kinase | Tyrosine Kinase ZAP-70 | Tyrosine-protein kinase ZAP-70 | Tyrosine-protein kinase ZAP-70 (Syk) | Tyrosine-protein kinase ZAP-70 (ZAP70) | Tyrosine-protein kinase ZAP70 | ZAP70 | ZAP70_HUMAN | Zeta-chain (TCR) associated protein kinase 70kDa |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 69881.61 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ZAP-70 SH2 domain was expressed and purified from E. coli using O-phospho-L-tyrosine-agarose column. |
|---|
| Residue: | 619 |
|---|
| Sequence: | MPDPAAHLPFFYGSISRAEAEEHLKLAGMADGLFLLRQCLRSLGGYVLSLVHDVRFHHFP
IERQLNGTYAIAGGKAHCGPAELCEFYSRDPDGLPCNLRKPCNRPSGLEPQPGVFDCLRD
AMVRDYVRQTWKLEGEALEQAIISQAPQVEKLIATTAHERMPWYHSSLTREEAERKLYSG
AQTDGKFLLRPRKEQGTYALSLIYGKTVYHYLISQDKAGKYCIPEGTKFDTLWQLVEYLK
LKADGLIYCLKEACPNSSASNASGAAAPTLPAHPSTLTHPQRRIDTLNSDGYTPEPARIT
SPDKPRPMPMDTSVYESPYSDPEELKDKKLFLKRDNLLIADIELGCGNFGSVRQGVYRMR
KKQIDVAIKVLKQGTEKADTEEMMREAQIMHQLDNPYIVRLIGVCQAEALMLVMEMAGGG
PLHKFLVGKREEIPVSNVAELLHQVSMGMKYLEEKNFVHRDLAARNVLLVNRHYAKISDF
GLSKALGADDSYYTARSAGKWPLKWYAPECINFRKFSSRSDVWSYGVTMWEALSYGQKPY
KKMKGPEVMAFIEQGKRMECPPECPPELYALMSDCWIYKWEDRPDFLTVEQRMRACYYSL
ASKVEGPPGSTQKAEAACA
|
|
|
|---|
| BDBM50305126 |
|---|
| n/a |
|---|
| Name | BDBM50305126 |
|---|
| Synonyms: | 7-(2-(dimethylamino)ethoxy)-2-(4-phenoxyphenyl)-9,10-dihydro-4H-benzo[d]pyrazolo[1,5-a][1,3]diazepine-3-carboxamide | CHEMBL591325 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H29N5O3 |
|---|
| Mol. Mass. | 483.5616 |
|---|
| SMILES | CN(C)CCOc1ccc2Nc3c(C(N)=O)c(nn3CCc2c1)-c1ccc(Oc2ccccc2)cc1 |
|---|
| Structure | 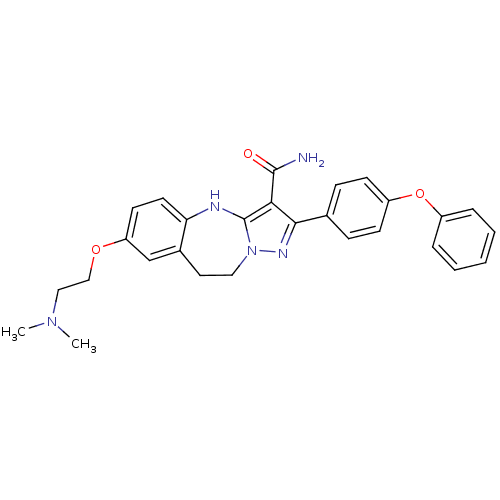
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Takayama, T; Umemiya, H; Amada, H; Yabuuchi, T; Koami, T; Shiozawa, F; Oka, Y; Takaoka, A; Yamaguchi, A; Endo, M; Sato, M Ring-fused pyrazole derivatives as potent inhibitors of lymphocyte-specific kinase (Lck): Structure, synthesis, and SAR. Bioorg Med Chem Lett20:112-6 (2010) [PubMed] Article
Takayama, T; Umemiya, H; Amada, H; Yabuuchi, T; Koami, T; Shiozawa, F; Oka, Y; Takaoka, A; Yamaguchi, A; Endo, M; Sato, M Ring-fused pyrazole derivatives as potent inhibitors of lymphocyte-specific kinase (Lck): Structure, synthesis, and SAR. Bioorg Med Chem Lett20:112-6 (2010) [PubMed] Article