

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cannabinoid receptor 1 | ||
| Ligand | BDBM50306012 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_605683 (CHEMBL1071213) | ||
| Ki | >10000±n/a nM | ||
| Citation |  Gilbert, EJ; Zhou, G; Wong, MK; Tong, L; Shankar, BB; Huang, C; Kelly, J; Lavey, BJ; McCombie, SW; Chen, L; Rizvi, R; Dong, Y; Shu, Y; Kozlowski, JA; Shih, NY; Hipkin, RW; Gonsiorek, W; Malikzay, A; Lunn, CA; Favreau, L; Lundell, DJ Non-aromatic A-ring replacement in the triaryl bis-sulfone CB2 receptor inhibitors. Bioorg Med Chem Lett20:608-11 (2010) [PubMed] Article Gilbert, EJ; Zhou, G; Wong, MK; Tong, L; Shankar, BB; Huang, C; Kelly, J; Lavey, BJ; McCombie, SW; Chen, L; Rizvi, R; Dong, Y; Shu, Y; Kozlowski, JA; Shih, NY; Hipkin, RW; Gonsiorek, W; Malikzay, A; Lunn, CA; Favreau, L; Lundell, DJ Non-aromatic A-ring replacement in the triaryl bis-sulfone CB2 receptor inhibitors. Bioorg Med Chem Lett20:608-11 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cannabinoid receptor 1 | |||
| Name: | Cannabinoid receptor 1 | ||
| Synonyms: | CANN6 | CANNABINOID CB1 | CB-R | CB1 | CNR | CNR1 | CNR1_HUMAN | Cannabinoid CB1 receptor | Cannabinoid receptor | Cannabinoid receptor 1 (CB1) | Cannabinoid receptor 1 (brain) | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 52868.96 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P21554 | ||
| Residue: | 472 | ||
| Sequence: |
| ||
| BDBM50306012 | |||
| n/a | |||
| Name | BDBM50306012 | ||
| Synonyms: | CHEMBL604683 | trans-4-chloro-2-(2-fluorophenylsulfonyl)-N-((1r,4r)-4-((trifluoromethylsulfonamido)methyl)cyclohexyl)benzenesulfonamide | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H21ClF4N2O6S3 | ||
| Mol. Mass. | 593.032 | ||
| SMILES | Fc1ccccc1S(=O)(=O)c1cc(Cl)ccc1S(=O)(=O)N[C@H]1CC[C@H](CNS(=O)(=O)C(F)(F)F)CC1 |r,wU:24.26,wD:21.22,(.36,-16.33,;-.98,-15.56,;-2.32,-16.33,;-3.65,-15.57,;-3.65,-14.02,;-2.32,-13.24,;-.98,-14.01,;.35,-13.23,;.74,-14.72,;1.83,-13.62,;.34,-11.7,;-.99,-10.94,;-1.01,-9.41,;-2.35,-8.64,;.33,-8.63,;1.67,-9.39,;1.67,-10.93,;3.01,-11.7,;2.6,-13.17,;4.33,-12.46,;3.76,-10.36,;5.3,-10.34,;6.08,-11.66,;7.61,-11.65,;8.37,-10.31,;9.9,-10.3,;10.66,-8.97,;12.19,-8.95,;12.17,-7.39,;13.52,-8.14,;12.99,-10.26,;14.53,-10.24,;12.24,-11.61,;13.77,-11.58,;7.59,-8.98,;6.04,-8.99,)| | ||
| Structure | 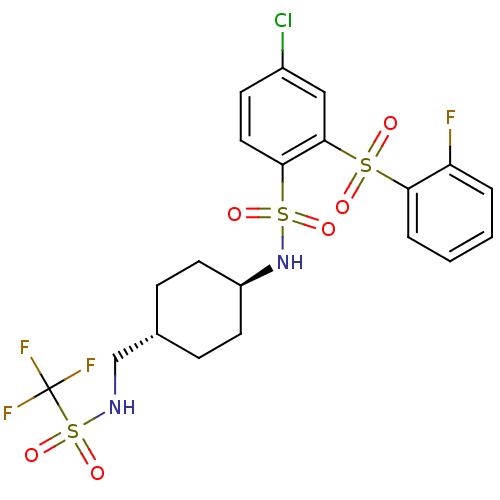
| ||