| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cAMP-dependent protein kinase catalytic subunit alpha |
|---|
| Ligand | BDBM50311409 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_622265 (CHEMBL1110410) |
|---|
| IC50 | 5.5±n/a nM |
|---|
| Citation |  Enkvist, E; Kriisa, M; Roben, M; Kadak, G; Raidaru, G; Uri, A Effect of the structure of adenosine mimic of bisubstrate-analog inhibitors on their activity towards basophilic protein kinases. Bioorg Med Chem Lett19:6098-101 (2009) [PubMed] Article Enkvist, E; Kriisa, M; Roben, M; Kadak, G; Raidaru, G; Uri, A Effect of the structure of adenosine mimic of bisubstrate-analog inhibitors on their activity towards basophilic protein kinases. Bioorg Med Chem Lett19:6098-101 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| cAMP-dependent protein kinase catalytic subunit alpha |
|---|
| Name: | cAMP-dependent protein kinase catalytic subunit alpha |
|---|
| Synonyms: | KAPCA_HUMAN | PKA C-alpha | PKACA | PRKACA | cAMP-dependent protein kinase (PKA) | cAMP-dependent protein kinase catalytic (PKA) | cAMP-dependent protein kinase catalytic subunit alpha | cAMP-dependent protein kinase catalytic subunit alpha (PKA) | cAMP-dependent protein kinase catalytic subunit alpha (PKACA) | cAMP-dependent protein kinase catalytic subunit alpha (PKAc) | cAMP-dependent protein kinase catalytic subunit alpha isoform 1 | cAMP-dependent protein kinase, alpha-catalytic subunit |
|---|
| Type: | Enzyme Catalytic Subunit |
|---|
| Mol. Mass.: | 40598.73 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 351 |
|---|
| Sequence: | MGNAAAAKKGSEQESVKEFLAKAKEDFLKKWESPAQNTAHLDQFERIKTLGTGSFGRVML
VKHKETGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMV
MEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGY
IQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFF
ADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFAT
TDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVSINEKCGKEFSEF
|
|
|
|---|
| BDBM50311409 |
|---|
| n/a |
|---|
| Name | BDBM50311409 |
|---|
| Synonyms: | CHEMBL1077375 | N-((6R,9R,12R,15R,18R,21R,31R)-1-amino-31-(4-aminobutyl)-6-carbamoyl-9,12,15,18,21-pentakis(3-guanidinopropyl)-1-imino-8,11,14,17,20,23,30,33-octaoxo-2,7,10,13,16,19,22,29,32-nonaazaoctatriacontan-38-yl)-5-(2-aminopyrimidin-4-yl)thiophene-2-carboxamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C63H114N32O10S |
|---|
| Mol. Mass. | 1511.853 |
|---|
| SMILES | [#7]-[#6]-[#6]-[#6]-[#6]-[#6@@H](-[#7]-[#6](=O)-[#6]-[#6]-[#6]-[#6]-[#6]-[#7]-[#6](=O)-c1ccc(s1)-c1ccnc(-[#7])n1)-[#6](=O)-[#7]-[#6]-[#6]-[#6]-[#6]-[#6]-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](-[#7])=O |r| |
|---|
| Structure | 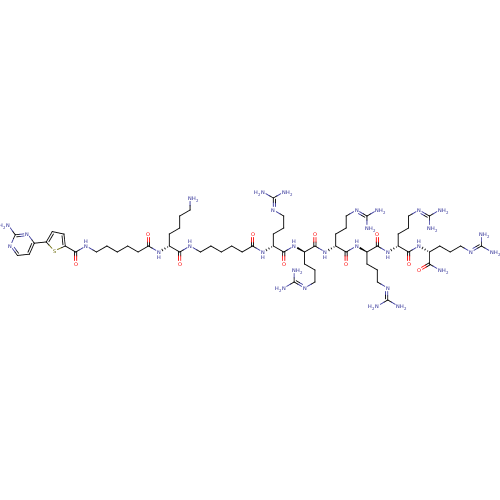
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Enkvist, E; Kriisa, M; Roben, M; Kadak, G; Raidaru, G; Uri, A Effect of the structure of adenosine mimic of bisubstrate-analog inhibitors on their activity towards basophilic protein kinases. Bioorg Med Chem Lett19:6098-101 (2009) [PubMed] Article
Enkvist, E; Kriisa, M; Roben, M; Kadak, G; Raidaru, G; Uri, A Effect of the structure of adenosine mimic of bisubstrate-analog inhibitors on their activity towards basophilic protein kinases. Bioorg Med Chem Lett19:6098-101 (2009) [PubMed] Article