| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Sphingosine 1-phosphate receptor 2 |
|---|
| Ligand | BDBM50158348 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_632508 (CHEMBL1111156) |
|---|
| EC50 | 44±n/a nM |
|---|
| Citation |  Bolli, MH; Abele, S; Binkert, C; Bravo, R; Buchmann, S; Bur, D; Gatfield, J; Hess, P; Kohl, C; Mangold, C; Mathys, B; Menyhart, K; Müller, C; Nayler, O; Scherz, M; Schmidt, G; Sippel, V; Steiner, B; Strasser, D; Treiber, A; Weller, T 2-imino-thiazolidin-4-one derivatives as potent, orally active S1P1 receptor agonists. J Med Chem53:4198-211 (2010) [PubMed] Article Bolli, MH; Abele, S; Binkert, C; Bravo, R; Buchmann, S; Bur, D; Gatfield, J; Hess, P; Kohl, C; Mangold, C; Mathys, B; Menyhart, K; Müller, C; Nayler, O; Scherz, M; Schmidt, G; Sippel, V; Steiner, B; Strasser, D; Treiber, A; Weller, T 2-imino-thiazolidin-4-one derivatives as potent, orally active S1P1 receptor agonists. J Med Chem53:4198-211 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Sphingosine 1-phosphate receptor 2 |
|---|
| Name: | Sphingosine 1-phosphate receptor 2 |
|---|
| Synonyms: | EDG5 | S1P2 | S1PR2 | S1PR2_HUMAN | Sphingosine 1-phosphate receptor | Sphingosine 1-phosphate receptor Edg-5 | Sphingosine-1-phosphate receptor 2 | ndothelial differentiation G-protein coupled receptor 5 |
|---|
| Type: | G Protein-Coupled Receptor (GPCR) |
|---|
| Mol. Mass.: | 38883.16 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Membranes isolated from S1P2-transfected CHO cells were used in ligand binding assay. |
|---|
| Residue: | 353 |
|---|
| Sequence: | MGSLYSEYLNPNKVQEHYNYTKETLETQETTSRQVASAFIVILCCAIVVENLLVLIAVAR
NSKFHSAMYLFLGNLAASDLLAGVAFVANTLLSGSVTLRLTPVQWFAREGSAFITLSASV
FSLLAIAIERHVAIAKVKLYGSDKSCRMLLLIGASWLISLVLGGLPILGWNCLGHLEACS
TVLPLYAKHYVLCVVTIFSIILLAIVALYVRIYCVVRSSHADMAAPQTLALLKTVTIVLG
VFIVCWLPAFSILLLDYACPVHSCPILYKAHYFFAVSTLNSLLNPVIYTWRSRDLRREVL
RPLQCWRPGVGVQGRRRGGTPGHHLLPLRSSSSLERGMHMPTSPTFLEGNTVV
|
|
|
|---|
| BDBM50158348 |
|---|
| n/a |
|---|
| Name | BDBM50158348 |
|---|
| Synonyms: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate | (2S,3R,4E)-2-amino-4-octadecene-1,3-diol 1-(dihydrogen phosphate) | CHEMBL225155 | US10676467, Compound S1P | US11584726, Example S1P | sphingosine 1-phosphate | sphingosine-1-phosphate |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H38NO5P |
|---|
| Mol. Mass. | 379.4718 |
|---|
| SMILES | CCCCCCCCCCCCC\C=C\[C@@H](O)[C@@H](N)COP(O)(O)=O |r| |
|---|
| Structure | 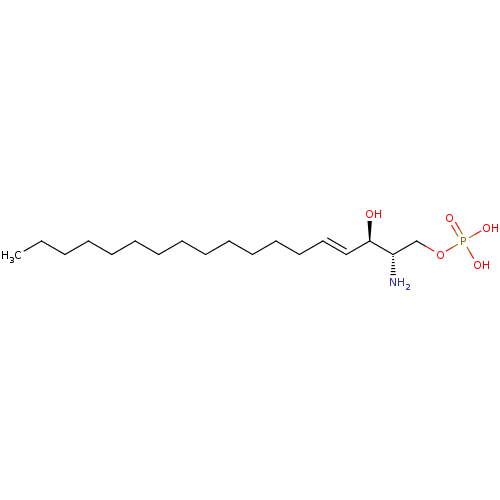
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Bolli, MH; Abele, S; Binkert, C; Bravo, R; Buchmann, S; Bur, D; Gatfield, J; Hess, P; Kohl, C; Mangold, C; Mathys, B; Menyhart, K; Müller, C; Nayler, O; Scherz, M; Schmidt, G; Sippel, V; Steiner, B; Strasser, D; Treiber, A; Weller, T 2-imino-thiazolidin-4-one derivatives as potent, orally active S1P1 receptor agonists. J Med Chem53:4198-211 (2010) [PubMed] Article
Bolli, MH; Abele, S; Binkert, C; Bravo, R; Buchmann, S; Bur, D; Gatfield, J; Hess, P; Kohl, C; Mangold, C; Mathys, B; Menyhart, K; Müller, C; Nayler, O; Scherz, M; Schmidt, G; Sippel, V; Steiner, B; Strasser, D; Treiber, A; Weller, T 2-imino-thiazolidin-4-one derivatives as potent, orally active S1P1 receptor agonists. J Med Chem53:4198-211 (2010) [PubMed] Article