

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Corticotropin-releasing factor receptor 1 | ||
| Ligand | BDBM50319907 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_635026 (CHEMBL1118577) | ||
| Ki | 9.1±n/a nM | ||
| Citation |  Zuev, D; Vrudhula, VM; Michne, JA; Dasgupta, B; Pin, SS; Huang, XS; Wu, D; Gao, Q; Zhang, J; Taber, MT; Macor, JE; Dubowchik, GM Discovery of 6-chloro-2-trifluoromethyl-7-aryl-7H-imidazo[1,2-a]imidazol-3-ylmethylamines, a novel class of corticotropin-releasing factor receptor type 1 (CRF1R) antagonists. Bioorg Med Chem Lett20:3669-74 (2010) [PubMed] Article Zuev, D; Vrudhula, VM; Michne, JA; Dasgupta, B; Pin, SS; Huang, XS; Wu, D; Gao, Q; Zhang, J; Taber, MT; Macor, JE; Dubowchik, GM Discovery of 6-chloro-2-trifluoromethyl-7-aryl-7H-imidazo[1,2-a]imidazol-3-ylmethylamines, a novel class of corticotropin-releasing factor receptor type 1 (CRF1R) antagonists. Bioorg Med Chem Lett20:3669-74 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Corticotropin-releasing factor receptor 1 | |||
| Name: | Corticotropin-releasing factor receptor 1 | ||
| Synonyms: | CRF-R | CRF-R2 Alpha | CRF1 | CRFR | CRFR1 | CRFR1_HUMAN | CRH-R 1 | CRHR | CRHR1 | Corticotropin releasing factor receptor 1 | Corticotropin-releasing factor receptor 1 (CRF-1) | Corticotropin-releasing factor receptor 1 (CRF1) | Corticotropin-releasing hormone receptor 1 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 50744.31 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P34998 | ||
| Residue: | 444 | ||
| Sequence: |
| ||
| BDBM50319907 | |||
| n/a | |||
| Name | BDBM50319907 | ||
| Synonyms: | CHEMBL1086468 | N-((2-chloro-1-mesityl-6-(trifluoromethyl)-1H-imidazo[1,2-a]imidazol-5-yl)methyl)-N-(cyclopropylmethyl)propan-1-amine | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H28ClF3N4 | ||
| Mol. Mass. | 452.943 | ||
| SMILES | CCCN(CC1CC1)Cc1c(nc2n(c(Cl)cn12)-c1c(C)cc(C)cc1C)C(F)(F)F |(-1.4,.94,;-2.94,.96,;-3.69,2.31,;-5.23,2.33,;-5.98,3.67,;-5.19,5,;-3.84,5.75,;-5.17,6.53,;-6.02,1.01,;-5.57,-.47,;-6.51,-1.7,;-5.63,-2.97,;-4.15,-2.52,;-2.68,-3.03,;-1.75,-1.8,;-.21,-1.83,;-2.63,-.53,;-4.11,-.98,;-1.92,-4.37,;-.39,-4.36,;.38,-3.03,;.38,-5.69,;-.39,-7.03,;.37,-8.37,;-1.93,-7.03,;-2.7,-5.69,;-4.24,-5.68,;-8.05,-1.67,;-8.79,-.32,;-8.85,-2.99,;-9.6,-1.66,)| | ||
| Structure | 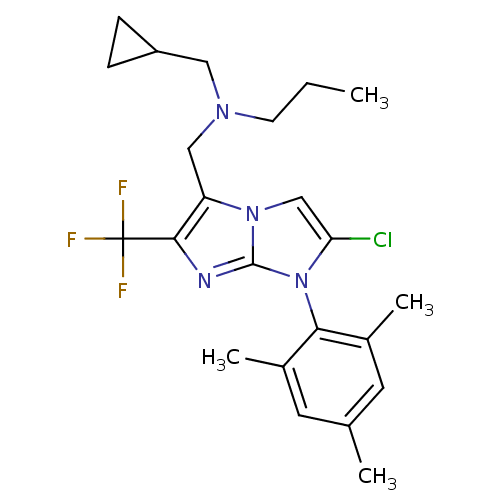
| ||