| Synonyms: | PI3-kinase p110 subunit gamma | PI3-kinase subunit p120-gamma | PI3Kgamma | PIK3CG | PK3CG_HUMAN | Phosphatidylinositol 4,5-biphosphate 3-kinase catalytic subunit gamma (PIK3CG) | Phosphatidylinositol 4,5-bisphosphate 3-kinase (PI3K) | Phosphatidylinositol 4,5-bisphosphate 3-kinase 110 kDa catalytic subunit gamma (PI3K gamma) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma (PI3Kgamma) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (PI3K gamma) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (PI3K) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (PI3Kgamma) | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | Phosphoinositide 3-Kinase (PI3K), gamma Chain A | Phosphoinositide 3-kinases gamma (PI3K gamma) | Phosphoinositide-3-kinase (PI3K gamma) | p120-PI3K |
|---|
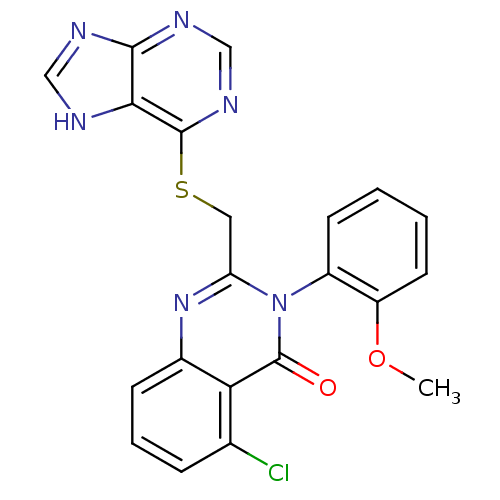
| SMILES | COc1ccccc1-n1c(CSc2ncnc3nc[nH]c23)nc2cccc(Cl)c2c1=O |(19.17,3.9,;19.17,2.36,;20.51,1.59,;21.84,2.36,;23.18,1.61,;23.18,.05,;21.86,-.72,;20.52,.04,;19.19,-.72,;19.2,-2.28,;20.54,-3.05,;20.54,-4.59,;21.87,-5.36,;21.86,-6.89,;23.22,-7.65,;24.55,-6.88,;24.53,-5.33,;25.67,-4.3,;25.02,-2.9,;23.5,-3.07,;23.19,-4.57,;17.86,-3.06,;16.53,-2.29,;15.19,-3.06,;13.86,-2.29,;13.85,-.75,;15.18,.02,;15.18,1.56,;16.52,-.75,;17.85,.03,;17.85,1.57,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Berndt, A; Miller, S; Williams, O; Le, DD; Houseman, BT; Pacold, JI; Gorrec, F; Hon, WC; Liu, Y; Rommel, C; Gaillard, P; Rückle, T; Schwarz, MK; Shokat, KM; Shaw, JP; Williams, RL The p110 delta structure: mechanisms for selectivity and potency of new PI(3)K inhibitors. Nat Chem Biol6:117-24 (2010) [PubMed] Article
Berndt, A; Miller, S; Williams, O; Le, DD; Houseman, BT; Pacold, JI; Gorrec, F; Hon, WC; Liu, Y; Rommel, C; Gaillard, P; Rückle, T; Schwarz, MK; Shokat, KM; Shaw, JP; Williams, RL The p110 delta structure: mechanisms for selectivity and potency of new PI(3)K inhibitors. Nat Chem Biol6:117-24 (2010) [PubMed] Article